Protein stability is critical to the success or failure of the development of a biopharmaceutical. Protein stability is an important parameter during production, manufacturing, formulation, long term storage, delivery to patient, and efficacy. Highly stable proteins will likely have fewer issues during the manufacturing process, are more cost-effective to produce, and will have a better chance of remaining functional during formulation and storage without chemical alteration or aggregation. In the “Quality by Design” (QbD) approach for biopharmaceutical development, stability characterization is part of the assessment of the 'developability' or 'drugability' of a potential drug candidate, as well as during process development and manufacturing. Stability data is also incorporated in higher order structure (HOS) characterization and 'fingerprinting' used for manufacturing support, biocomparability and biosimilarity. Protein HOS characterization is also increasingly expected in regulatory submissions for new biopharmaceutical drugs and biosimilars.
Due to the complex nature of proteins, biophysical tools are important in the complete characterization of a biopharmaceutical product. There are several biophysical tools used to assess protein stability, including (but not limited to) circular dichroism (CD), dynamic and static light scattering (DLS and SLS), size exclusion chromatography–multi-angle light scattering (SEC-MALS), Fourier transform infrared spectroscopy (FTIR), analytical ultrafiltration (AUC), size exclusion chromatography (SEC), differential scanning fluorescence (DSF), intrinsic fluorescence (IF) and differential scanning calorimetry (DSC).
While all of these biophysical assays play an important role in biopharmaceutical development, characterizing thermal stability by DSC is critical. In a 2015 article about biophysical techniques for monoclonal antibody higher order structure characterization, Gokarn et al. stated: "DSC remains as an unparalleled technique to assess the thermodynamic stability of proteins in a given buffer condition”[1].
The focus of this whitepaper is on the use of DSC to characterize the thermal stability of protein biopharmaceuticals (primarily antibodies), and as a HOS characterization tool for the comparability of biopharmaceuticals (batch-to-batch comparison, effect of process changes, etc), and for the development of biosimilars.
DSC is a microcalorimetry technique that is used to characterize the thermal and conformational stability of proteins, nucleic acids, lipids, and other biopolymers[2-7]. DSC measures heat capacity as a function of temperature. The DSC instruments used for protein characterization described in this whitepaper are “power compensation” instruments, with the biopolymer in solution, placed in a fixed-in-place sample cell, and a matched reference cell filled with buffer. The heat capacity (Cp) signal from a sample cell is compared with that from the reference cell. As the temperature of the cells is increased, the temperature difference between the reference and sample cells is continuously measured and calibrated to power units. DSC is a 'forced degradation' assay - as the protein is exposed to increasing temperature, it begins to unfold and the Cp of the protein increases (Figure 1).
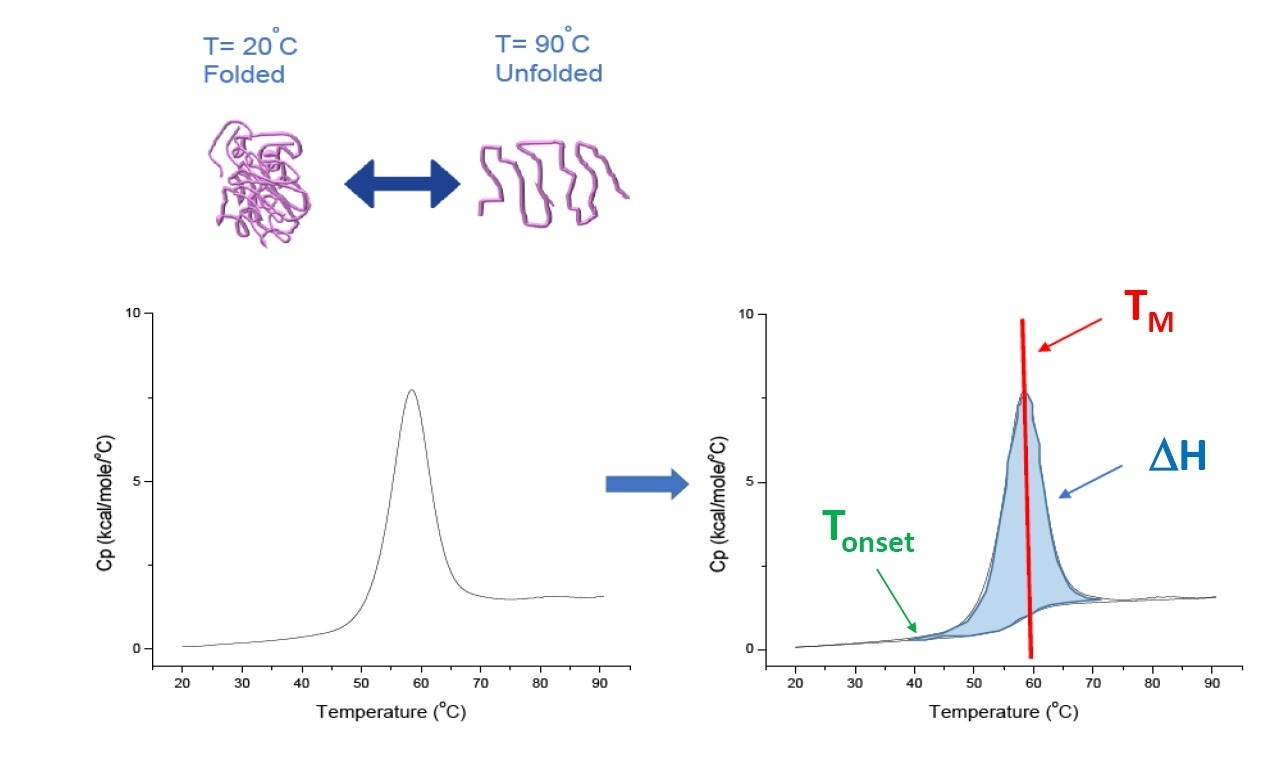
Figure 1: How DSC works. The heat capacity (Cp) changes as a protein thermally denatures. The DSC experiment starts at a temperature at which the protein is primarily folded in its native conformation. With increasing temperature, at some point the protein will begin to unfold/denature (Tonset) and the Cp increases. At the temperature where 50% of the protein is in its native conformation, and 50% is denatured, the Cp will reach its maximum value - this is the thermal transition midpoint or TM. Above the TM the protein will be primarily denatured, and at the end of the DSC experiment all of the protein will be in its unfolded conformation. Experimental parameters for DSC include Tonset, TM, and the unfolding enthalpy (ΔH)
DSC directly measures the heat capacity change, without need for any fluorescence or other label or probe. For a protein which reversibly denatures, the thermal transition midpoint (the TM), also called the melting or denaturation temperature, is the temperature at which 50% of the protein is in its native (folded) conformation, and 50% is in its denatured conformation. The TM is seen as the 'peak' of a DSC thermogram.
TM is considered a good indication of thermal stability – the higher the TM, the more thermally stable the protein. Multi-domain proteins (like antibodies) typically have more than one peak on a DSC thermogram, so more than one TM can be determined for each domain (see Figure 2).
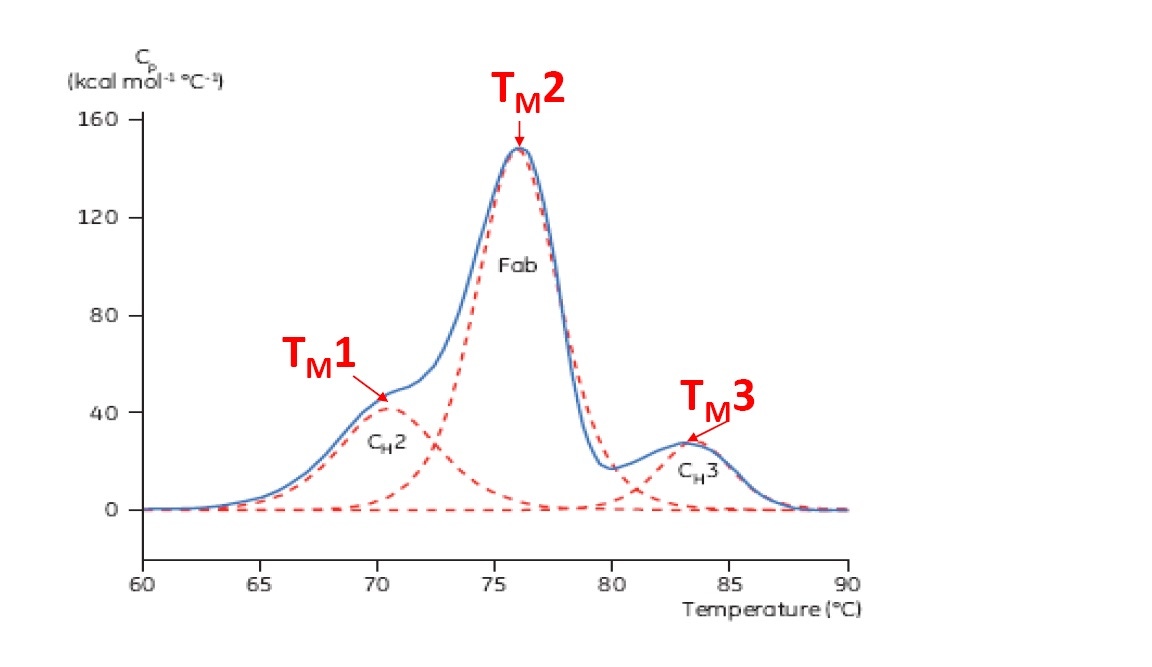
Figure 2: Representative DSC thermogram of a monoclonal antibody, with CH2, Fab, and CH3 domains identified. The dashed red lines are the deconvoluted peaks of each domain transition, with the three TMs indicated
DSC provides other useful parameters which can be used to characterize and rank-order protein stability, including the unfolding enthalpy (ΔH), which is measured by the area under the curve. Protein unfolding is endothermic, since energy input is needed to break the secondary non-covalent bonds that keep the protein folded. DSC also determines the Tonset (start of unfolding), ΔCp (heat capacity change of unfolding), and T1/2 (width at 1/2 the peak height, indicative of the shape of the unfolding thermogram). DSC analysis can include the determination of any combination of these parameters.
Most proteins denature irreversibly, and tend to aggregate or precipitate after thermal denaturation. The TM and other parameters from the DSC analysis of irreversibly denatured proteins are not true thermodynamic parameters. However, the rank-ordering of TM from the DSC of an irreversible protein denaturation remains very useful for qualitative parameter stability screening.
Malvern Instruments offers the MicroCal VP-Capillary DSC system[8,9] which is an automated DSC designed for TM screening and thermodynamic characterization of proteins and biopolymers in solution.
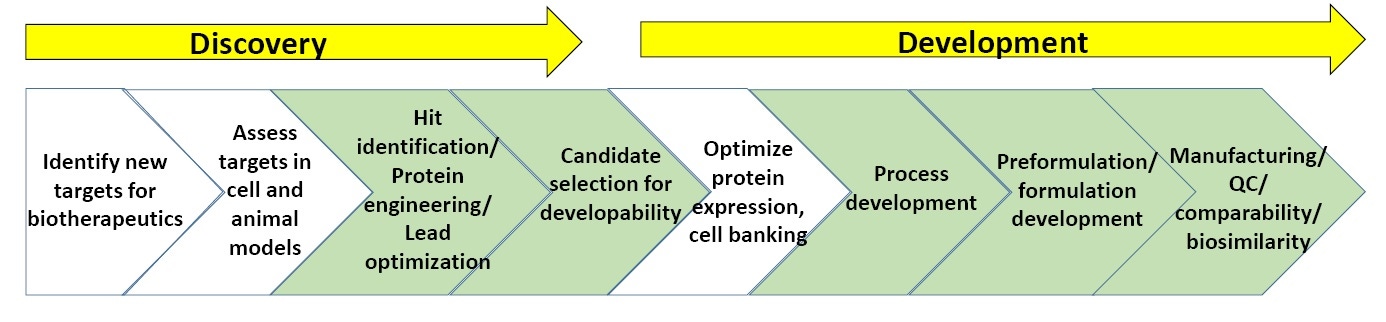
Figure 3: General schematic of processes within biopharmaceutical discovery and development
Figure 3 shows a general schematic of biopharmaceutical discovery and development processes. The sections in green are the points at which biophysical characterization, including stability assays, is most commonly utilized (at the end of this whitepaper is a list of suggested reading on biopharmaceutical discovery and development).
In the development of a desirable biopharmaceutical, scientists initially look for biomolecules that already demonstrate high stability during candidate selection, and may need to introduce increased stability via protein engineering. During purification, the protein is removed from conditions where it is stable, correctly folded, and active, so it is very important to use buffers, additives, purification, and storage conditions that can keep the protein as stable as possible throughout this process.
Formulated subcutaneous (SC) protein drugs must be stable and unaffected at very high protein concentrations (sometimes around 100 mg/mL) within their container closure, (e.g., vial or prefilled syringe) for several years. When protein molecules are exposed to stresses like heat, chemicals, pH changes, pressure, mixing, and high concentration, any and all of which may occur during biopharmaceutical production and formulation, the protein conformation can favor the denatured (unfolded) protein. Proteins in solution are also susceptible to modifications like deamidation and oxidation that will also lead to denatured, inactive proteins. In the case of a protein biopharmaceutical, denaturation and other modifications could result in the formation of aggregates that have reduced efficacy or are nonfunctional as a drug. More significantly, protein aggregation can result in a potentially fatal immunogenic response in a patient. Using a stable protein as the basis for a biopharmaceutical will lead to more cost-effective production, and a successful, effective and safe drug product.
The sequence of amino acids, or a protein's primary (1°) structure, is the most basic component of the polypeptide chain and protein structure. Beyond this, it is important to understand and characterize the protein’s three-dimensional structure, also called higher order structure (HOS). There are three levels of protein HOS: secondary (2°), referring to the local folding patterns of a protein’s primary structure, including α-helix, β-sheet, turns, and random coils; tertiary (3°), the final 3D structure of a protein, arising from an array of secondary structural elements; quaternary (4°), which describes structures that involve the interaction of two or more identical or different polypeptide chains.
DSC is a measure of the conformational stability and changes in tertiary and quaternary structure that occur when a protein is thermally denatured, as well as providing an indication of how intrinsic and extrinsic factors affect a protein's stability. DSC is considered the best and most quantitative assay for thermal stability used in the characterization of biopharmaceutical proteins, as a predictor of long-term stability[1,10-14]. TM from DSC is a parameter frequently used to rank-order stability in candidate selection (developability), formulation screening, and process development, with more stable proteins having a higher TM. Enthalpy (ΔH), Tonset, T1/2, and ΔCp from DSC are also used in rank-ordering stability, validation of DSC data, quantitative analysis of protein unfolding, and higher order structure 'fingerprinting'[10-14].
DSC analysis of a protein in defined solution conditions is quantitative and reproducible if the protein is the same or highly similar (Figure 4). That is, the DSC thermograms will have a reproducible pattern, and parameters (including TM, ΔH and Tonset) will be within an accepted range[12-14]. If compared thermograms are different and the DSC fit parameters change, it suggests that there has been an event such as protein misfolding, degradation, aggregation, change in solvent, post-translational modification, or other higher order structure change, affecting the conformational stability.
Reproducible and quantitative data make DSC a key assay in comparability and HOS analysis for product evaluation during manufacturing (including batch-to-batch and site-to-site comparability), the comparison of protein variants and modified products (including structural changes due to glycosylation and oxidation), and biosimilarity. DSC data are also used in regulatory support documents as a HOS characterization for new drug and biosimilar submissions. In a survey of scientists in the biopharmaceutical industry, DSC was ranked as a "very useful" to "extremely useful" HOS biophysical tool for candidate selection, formulation development, product characterization, comparability, process development, and biosimilarity[15].
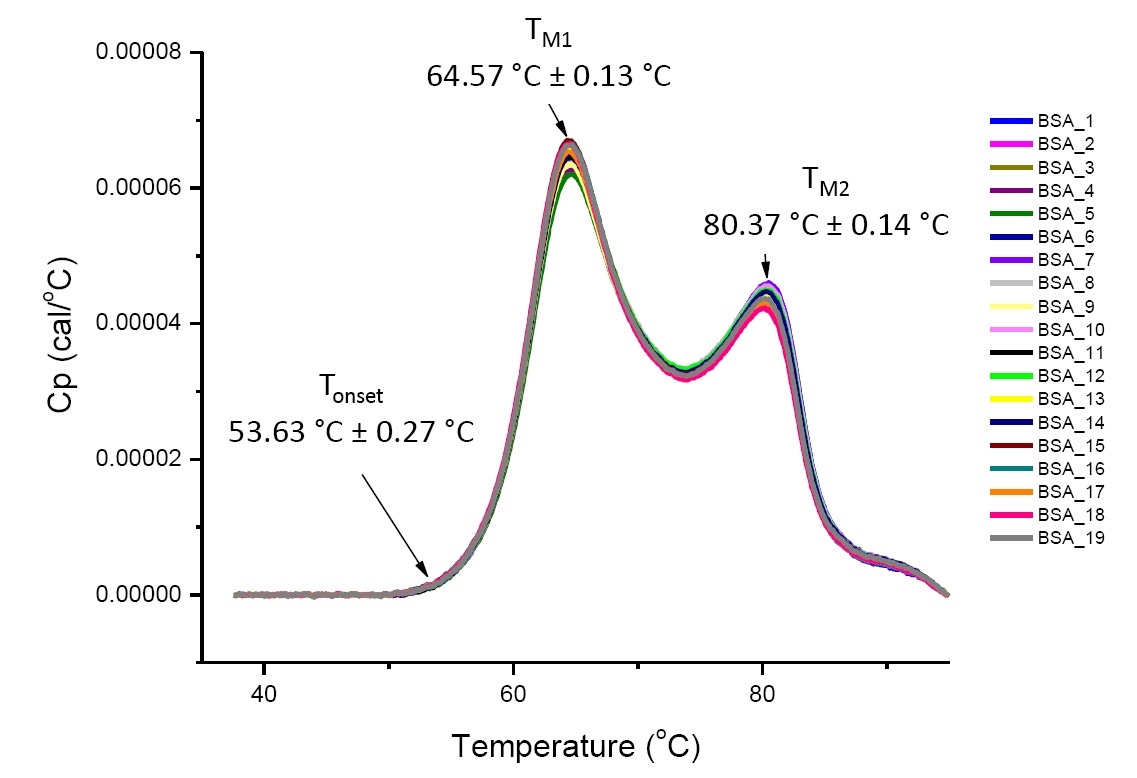
Figure 4: Nineteen DSC thermograms of bovine serum albumin (Sigma A1933, chromatographically-purified) in PBS. DSC data shown after scan rate normalization, buffer-buffer subtraction, and integration baseline subtraction. Mean and standard deviations of Tonset, TM1, and TM2 are shown
For multi-domain proteins like antibodies, DSC thermograms have more than one unfolding transition (as seen in Figure 2). DSC is able to characterize and quantitate the different domains, and determine the individual TMs for each transition. The TM values are represented by the thermogram peak(s) and can be simply determined from the DSC data, without the need for complex data analysis. Alternative biophysical assays which can determine TM, such as CD, IF and DSF, may only detect the first TM (occurring at the lowest temperature), or the 'most dominant' TM for multi-domain proteins. Extraction of more than one TM from spectroscopic and fluorescent data requires complex data fitting and may not be reproducible.
DSC does require more protein sample per scan, and can be lower-throughput, compared to other TM screening assays. If sample is limited, a good solution would be to perform an initial TM rank-ordering with DSF or IF, then choose several samples to validate TM using DSC. It is important to validate the TM results with DSC, and not rely solely upon fluorescence or spectroscopy for the calculation of TM to assess stability. It is common in fluorescence-based assays to see artifacts which interfere with the output, and the TM results may appear to shift to a higher (or lower) value due to these artifacts. Some proteins and buffer conditions are not compatible with fluorescence, which can make the calculation of TM differences very complicated. Finally, fluorescence and spectroscopy dare not able to determine the calorimetric enthalpy and other thermodynamic parameters, while DSC is able to provide all of this information as a thermal stability 'data suite'.
DSC is considered by the biopharmaceutical industry as the “gold standard” thermal stability assay because the technique:
Measures heat changes associated with protein unfolding
Is a direct measurement of protein unfolding, so no label, probe or tag is needed. This means that there are no potential detection artifacts which are commonly found in fluorescence or other spectroscopic assays
Measures native proteins in solution, and can be used with practically all buffers and additives which are in common use within biopharmaceutical purification and formulation development. Some of these buffers and additives are not compatible with fluorescence or spectroscopy
Is easy to use for experimental set-up
Has high precision temperature control, with a working temperature range of up to 130°C, so can detect most high TM transitions. Other TM screening assays are able to heat samples up to only 100°C (or lower)
Is a 'forced degradation' assay, so does not require storage of the protein in buffer prior to analysis. SEC-HPLC and DLS often require incubating samples in buffer at elevated temperature to detect changes in conformation
Has simple data output and integrated data analysis software
Can be used to resolve separate unfolding transitions and characterize multi-domain proteins and protein complexes, as well as simple single-domain proteins
Is information-rich, providing thermodynamic data as well as conformational stability and TM determination
Can be used as a primary assay for biotherapeutic thermal stability characterization, and can also be used with other biophysical screening tools which are orthogonal or complementary, and/or to validate other data
Is available with high-throughput automation (MicroCal VP-Capillary DSC system) for rapid screening of thermal stability
Biopharmaceutical comparability
Monoclonal antibodies and other biopharmaceutical drugs are complex proteins expressed by mammalian or bacterial cells. Each protein is unique and requires extensive research, characterization and optimization in order to transform it into a drug. As part of discovery and early development, the desired properties of the protein drug are defined, including purity, potency, and dosage. Biochemical, biophysical, and biological assays are developed to study these characteristics, and are optimized throughout product development.
Detailed protein characterization and HOS (including stability) is carried out for each process to define the protein's critical quality attributes and critical process parameters, needed to control and validate processes throughout the entire product life cycle. In the “Quality by Design” (QbD) approach to biopharmaceutical development, critical quality attributes and stability characterization are part of the assessment of the drug during process development and manufacturing support[16,17].
During the processing of a protein drug, the protein is exposed to different conditions, including:
Different solutions, including varying buffer, pH, and salts
Formulation additives (excipients)
Range of protein concentrations
Repeated freeze/thaw cycles, increased pressures and mixing/agitation
Different material surfaces the protein comes in contact with (pumps, ultrafiltration/diafiltration membranes, chromatography media, etc.)
Exposure to oxidants, proteases, cell growth media, product variants, etc.
Any of these conditions can disrupt the forces and interactions that keep a protein in its native folded conformation, resulting in denatured or inactive protein. Chemical denaturation due to oxidation, deamidation, changes in glycosylation or other post-translational modifications, may also take place.
Denatured proteins often form aggregates, which is a key issue for biopharmaceutical development. Aggregates are associated states of the protein monomer, and aggregate formation can be reversible or irreversible, ranging in size from a protein dimer to large particles that are visible to the naked eye. As a minimum, protein aggregation reduces the drugs efficacy and potency, and will increase the cost of production. The greater concern about protein aggregates and particles is their potential to give rise to immunogenic responses, which in extreme cases may result in patient death. Because of this issue, protein aggregation has received considerable scrutiny within the biopharmaceutical industry. This includes particle/aggregation detection, quantification and characterization during production and formulation, and a search for solutions to reduce or eliminate protein aggregation in biopharmaceutical products.
It is important to demonstrate that the manufactured protein drug is comparable in structure, stability, size distribution, and biochemical and functional assays to:
The same protein manufactured in previous lots and also the reference protein lot
The same protein manufactured at different sites
The same protein manufactured using a modified upstream or downstream process
Scale-up of upstream or downstream process
The same protein in a different formulation
Biochemical, biophysical, HOS, and functional assays for comparability (or biocomparability) are designed to demonstrate that the manufactured protein product is 'highly similar' in the designated critical quality attributes, compared to a reference protein. It is essential that the HOS of protein drug candidates is carefully analyzed at each stage of development.
Changes to the manufacturing process of a biopharmaceutical may occur both during process development and after drug approval. The reasons for such changes include:
Improvement of the manufacturing process (e.g. product yield, reduced cost)
Increase in scale
A manufacturing site change
Compliance and regulatory changes
Improvement of product stability
When significant changes are made to the manufacturing process, a comparability exercise to evaluate the impact of the change(s) on the critical quality attributes, safety and efficacy of the biopharmaceutical product should be conducted. The demonstration of comparability does not necessarily mean that the critical quality attributes of the pre-change and post-change product are identical, but rather that they are highly similar. Manufacturing changes should have no negative impact on product quality.
Comparability is a major concern for protein therapeutics, and is addressed by several international regulatory agencies [18,19,20]. No single analytical method can be used for comparability assessment of protein drugs. HOS and comparability studies include (but are not limited to): DSC, DLS, fluorescence, CD, AUC, SEC, Raman spectroscopy, Nanoparticle Tracking Analysis (NanoSight from Malvern Instruments), resonant mass measurement (Archimedes from Malvern Instruments), and microscopy. Protein bioactivity and efficacy are also monitored using bioassays, isothermal titration calorimetry (ITC) and surface plasmon resonance (SPR).
The information provided by each biophysical method is specific, and when assessed together, provides structural information for HOS characterization and comparability. The HOS 'map' helps ensure consistency in protein structure upon scale-up during process development cycle, lot-to-lot comparability, when processes transfer to new manufacturing sites, and when production is modified. Information from HOS characterization is also invaluable for evaluating biosimilars.
It is important to note that testing and establishing comparability during a manufacturing process (or change) is not the same as the testing performed during development of a biosimilar drug product – biosimilar development is a far more complex process. Since a biosimilar is not normally produced by the same company that developed the innovator (also called parental or reference product), biosimilar development requires 'reverse engineering' to establish new upstream and downstream processes. The biosimilar manufacturer has to match product quality (biosimilarity) within the narrow range of the innovator’s commercial product, requiring more biophysical and biochemical evidence to demonstrate biosimilarity than what is typically needed to show comparability during manufacturing.
Since DSC provides information on the thermal stability of a protein under different solvent conditions, and DSC results are reproducible (Figure 4), DSC is often included in comparability studies to show that the process does not result in a change in product stability, and that the manufactured products from different lots and/or manufacturing sites are highly similar.
Jiang and Nahri[21] reviewed several biophysical techniques for comparability during process development and manufacturing. They cited the use of DSC as one of the biophysical characterization tools used in comparability of a monoclonal antibody drug product expressed by two different cell lines (cell lines 1 and 2), which also experienced a change in the production process (processes 2pA and 2pB). The three resulting protein products were assessed by FTIR (to determine secondary structure), near-UV CD and intrinsic fluorescence (to compare tertiary structure) fluorescence and ANS binding (to compare surface hydrophobicity), DLS (to compare the hydrodynamic properties and size distribution of the proteins), and DSC (to compare thermal stability and solubility).
The results from this suite of biophysical assays suggested that the overall secondary and tertiary structures of the three samples were similar. The DSC results indicated that the thermal stability of the protein from cell line 2pB increased significantly compared with both the 2pA sample and the cell line 1 sample. The 2pA and cell line 1 samples also appeared more heterogeneous, according to DSC. Normally, 2 to 3 transitions are seen for antibodies (see Figure 2 for an example of a 'typical' antibody DSC thermogram). The presence of a higher number of overlapped thermal transitions, according to the DSC analysis of these samples, indicated sample heterogeneity. The DSC results suggested that the process change improved the homogeneity and stability of the monoclonal antibody of interest. Based on the biophysical, biochemical, and functional assay comparability data discussed in the article, cell line 2 and process 2pB were deemed to produce the more stable protein[21].
A second example from Jiang and Nahri[21] used DSC to look at the comparability of a protein product produced at different manufacturing sites. Protein Y was comprised of two monomeric polypeptide chains, disulfide-linked through the Fc region of the molecule. During the course of its development, Protein Y was manufactured at different sites. To verify that the protein present in these different lots was comparable in native conformation, secondary and tertiary structures, thermal stability, and size distribution, four different samples of Protein Y, representing three samples manufactured at different sites, and the reference protein standard, were analyzed by FTIR, far-UV CD, near-UV CD, fluorescence, DSC, and AUC.
The DSC scans of the reference standard and 3 samples from different manufacturing sites showed two thermal transitions for each sample, and all four DSC profiles were identical within experimental variability. This suggested that there was no difference in the thermal stability of the protein samples, and that they were all folded into the native conformation. The combined biophysical characterization results, including those produced by DSC, demonstrated that all four Protein Y samples were comparable, had the appropriate secondary and tertiary structure, and were homogeneous[21].
If the DSC thermograms had not been identical, it may have indicated a change in post-translational modification, or that chemical denaturation had occurred in the protein, related to the process or formulation. Arthur, et al.[22] demonstrated the sensitivity of DSC in the detection of HOS changes in stability arising from oxidation, a common chemical degradation pathway. If a biopharmaceutical product is subjected to oxidation during purification, or storage in the final formulation, it can negatively impact efficacy, and also increase the likelihood of protein aggregation formation. In this study, protein products from three structural classes were evaluated at multiple levels of oxidation. Each protein showed a linear decrease in TM as a function of methionine oxidation; the authors also observed differences in the rate of change in TM, as well as differences in domain TM stability, across and within structural classes[22]. In contrast, near-UV CD and fluorescence spectroscopy were far less sensitive to oxidation-induced conformational changes - DSC is a more appropriate structural characterization method for monitoring protein oxidation, compared with these spectroscopic methods[22]. For one protein in the study (IgG2B), changes in TM were detected by DSC preceding a loss in relative potency, demonstrating that DSC is a leading indicator of decreased antigen binding. Detectable changes in oxidized methionine by mass spectrometry (MS) occurred at oxidation levels below those with a detectable conformational or functional impact. Using TM shifts from DSC together with MS and potency methods, the relationship between a primary structural modification, changes in HOS and conformational stability, and functional impact can be reliably characterized.
Morar-Mitrica et al.[12] describe several case studies incorporating DSC in HOS and comparability studies of biopharmaceuticals, including the characterization of monoclonal antibody oxidation by observable TM shifts (similar to those described in [22]), reproducible DSC thermogram profiles, and the use of TM, T1/2, Tonset, and ∆H for comparability studies of biopharmaceuticals, and extended DSC characterization of glycosylated monoclonal antibodies with different levels of glycosylation (a common post-translational modification).
Shahrokl et al.[23] discuss how DSC is useful in comparability studies as part of the biological license application, mentioning DSC’s ease of use, and the simplicity of its data output. The authors describe how DSC was used for comparability of a 51 kDa glycoprotein before and after a change in its manufacturing process. The process change was implemented to increase product yield as well as remove animal-derived materials originating in the cell culture medium. Using DSC, the authors evaluated three lots of the glycoprotein which were produced using each manufacturing process, and observed that the DSC thermograms were superimposable and showed a single transition at 60.7 °C +/- 0.1 °C. The DSC results showed that the manufacturing change did not affect the conformational stability of the glycoprotein[23].
Lubiniecki et al.[24] evaluated the impact of manufacturing changes on antibody structure and function during the course of product development, by performing three comparability studies for two different IgG1 monoclonal antibody candidates (antibody A and antibody B). Two of the comparability studies incorporated DSC as one of the biochemical and biophysical analytical tools. One of the studies looked at process scale-up and transfer to the manufacturing site, together with the change from a lyophilized to a liquid dosage form. The DSC thermograms for the lyophilized and liquid formulations for both antibody A and antibody B showed good correlation[24]. DSC, used in conjunction with MS, CD, SEC, and other assays, showed that the manufacturing change did not result in the structure or function of the antibodies.
The third comparability study evaluated the antibody liquid formulation in prefilled syringes or vials[24]. Results from DSC, CD, SEC and other assays suggested similar molecular structure, biological activity, and degradation profiles. The one notable exception was a small (yet significant) increase in the levels of subvisible particles in prefilled syringes[24].
A biosimilar product (also called a “follow-on biologic” or “subsequent entry biologic”) is a biopharmaceutical that has been approved by a regulatory agency based on its high similarity to an previously-approved biological product, known as a reference product (also known as a parental or innovator product) . Biosimilars have been available globally for several years[25-29]. At the time this white paper was written (October 2016) the US FDA has approved four biosimilars, and the EMEA has approved 19 biosimilars.
Note that developing a biosimilar is not the same as developing a generic small molecule drug. Small molecule drugs are chemicals, and the synthesis of small molecule drugs is a controlled process. Since biopharmaceuticals are normally proteins. there is a certain degree of protein variation, even between different batches of the same product, due to the inherent variability of the biological expression system and the manufacturing process. These variations can include differences in post-translational modification and HOS. Because of proteins' high molecular weights, complex structures and natural variations, biosimilars are not as simple to produce as generics. A biosimilar is not an exact duplicate of the reference product, and it is for this reason that it is normally characterized as 'similar’ or 'highly similar' to the reference product.
The development of a biosimilar involves physicochemical, analytical and functional comparisons of the reference protein and the biosimilar. These assays are complemented by comparative non-clinical and clinical data to establish equivalent efficacy and safety. Before approval, it must be verified that the biosimilar has no clinically-meaningful differences in terms of safety and effectiveness to that of the reference product. Only minor differences in clinically-inactive components are allowable in biosimilar products. The biosimilar is expected to work in exactly the same way as the reference product for the same approved indications.
Regulatory agencies evaluate biosimilars based on their level of similarity to their reference products. Due to the complexity of biopharmaceuticals, it is not likely for two different manufacturers to produce two identical products, even if identical host expression systems, processes, and equivalent technologies are used. Therefore, biosimilar producers need to rely on comparability studies and analysis of HOS as discussed above[30,31]. The need for increased analytical information, and the desire for compressed timelines in biosimilar development demands the demonstratation of comparability to the reference molecule at every stage, particularly during manufacturing.
Biosimilars must be 'reverse-engineered' to match the reference product as closely as possible. The analytical characterization of a biosimilar includes primary and higher order structure (secondary, tertiary, and quaternary) assessment, biological activity, and analysis of product and process impurities. DSC is commonly used as a HOS biophysical assay to show that a biosimilar has a highly similar DSC profile 'fingerprint', and similar TM, Tonset, and other thermodynamic parameters, compared to the reference product.
Three of the four biosimilars approved by the US FDA (as of October 2016) included DSC as one of the HOS assays in the FDA filing:
Erelzi (from Sandoz, Inc.) is a biosimilar to Amgen’s Enbrel (etanercept)[32]
Inflectra (from Celltrion), called Remsima in other markets, is a biosimilar to Janssen’s Remicade (infliximab) [33,34]
Amjevita (from Amgen) is a biosimilar to Abbvie’s Humira (adalimumab)[35,36]
Sinha-Datta et al.[37] demonstrated the use of DSC (MicroCal VP-Capillary DSC) and surface plasmon resonance (SPR) as analytical tools to compare two therapeutic monoclonal antibodies (mAb1-i and mAb2-i) with their biosimilars (mAb1-B and mAb2-B1, B2, B3), based on thermal stability, kinetics and affinity. They observed that the biosimilars were highly similar to their reference samples, with respect to biofunction from SPR results. DSC was used as another confirmation of biosimilarity between the parental and biosimilar products. DSC analysis showed good structural similarity for the parental and biosimilar antibodies, with the primary TM at 84.1°C (for mAb1) and 72.8°C (for mAb2).
Information presented in this whitepaper demonstrates the importance and value of incorporating DSC as a biophysical stability and HOS assay for biopharmaceutical comparability and biosimilarity characterization. Using DSC results, along with other biophysical and biochemical assays, biopharmaceutical companies can make informed decisions on protein stability and comparability during production, ensure each batch of protein is highly similar to the reference lot, and that any process or manufacturing changes do not affect the conformational protein stability. DSC is also included as a HOS assay for biosimilar development, to help demonstrate that the biosimilar is 'highly similar' to the innovator.
Analytical Techniques for Biopharmaceutical Development, R. Rodriguez-Diaz, T. Wehr, S. Tuck (eds.), Taylor & Francis, New York, NY, USA (2005).
Biophysical Characterization of Proteins in Developing Biopharmaceuticals, D.J. Houde, S.A. Berkowitz (eds.), Elsevier, Amsterdam, Netherlands (2015).
Biophysical Methods for Biotherapeutics: Discovery and Development Applications, T.K. Das (ed.) John Wiley & Sons, Hoboken, NJ, USA (2014).
Biophysics for Therapeutic Protein Development, L.O. Nahri (ed.), Springer, New York, NY, USA (2013).
State-of-the-Art and Emerging Technologies for Therapeutic Monoclonal Antibody Characterization Volume 1. Monoclonal Antibody Therapeutics: Structure, Function, and Regulatory Space, J.E. Schiel, D.D. Davis, O.V. Borisov (eds.), ACS Symposium Series Vol 1176 (2014). doi: 10.1021/bk-2014-1176.
State-of-the-Art and Emerging Technologies for Therapeutic Monoclonal Antibody Characterization Volume 2. Biopharmaceutical Characterization: The NISTmAb Case Study, J.E. Schiel, D.D. Davis, O.V. Borisov (eds.), ACS Symposium Series Vol 1201 (2015). doi: 10.1021/bk-2015-1201.
State-of-the-Art and Emerging Technologies for Therapeutic Monoclonal Antibody Characterization Volume 3. Defining the Next Generation of Analytical and Biophysical Techniques, J.E. Schiel, D.D. Davis, O.V. Borisov (eds.), ACS Symposium Series Vol 1202 (2015) DOI: 10.1021/bk-2015-1202.
1. Gokarn, Y., Agarwal, S., Arthur, K., et al., Biophysical Techniques for Characterizing the Higher Order Structure and Interactions of Monoclonal Antibodies, in: State-of-the-Art and Emerging Technologies for Therapeutic Monoclonal Antibody Characterization Volume 2. Biopharmaceutical Characterization: The NISTmAb Case Study. J.E. Schiel, D.D. Davis, O.V. Borisov (eds.), ACS Symposium Series Vol 1201, American Chemical Society, Washington DC, USA, pages 285-327 (2015).
2. Cooper, A., Nutley, M. A., and Wadood, A., Differential Scanning Calorimetry, in: Protein-Ligand Interactions: Hydrodynamics and Calorimetry, A Practical Approach, S.E. Harding, B.Z. Choudry (eds). Oxford University Press, Oxford, UK, p. 287-318 (2001).
3. Malvern Instruments Whitepaper “Differential Scanning Calorimetry (DSC) Theory and Practice” http://www.malvern.com/en/support/resourcecenter/Whitepapers/WP140701-dsc-theory-and-practice.aspx.
4. Bruylants, G., Wouters, J., and Michaux, C., Current Med. Chem. 12, 2011-2020 (2005) doi: 10.2174/0929867054546564.
5. Jelesarov, I., and Bosshard. H.R., J. Mol. Recognit. 12, 3-18 (1999) doi: 10.1002/ (SICI)1099 1352(199901/02)12:1<3:AID-JMR441>3.0.CO; 2-6.
6. Choi, M.H., and Prenner, E.J., J. Pharm. Bioallied Sci. 3, 39-59 (2011) doi: 10.4103/0975-7406.76463.
7. Johnson, C.M., Arch. Biochem. Biophys. 531, 100-109 (2013) doi: 10.1016/j.abb.2012.09.008.
8. Plotnikov, V., Rochalski, A., Brandts, M., Brandts, J.F., Williston, S., Frasca, V., and Lin, L.N., Assay Drug Devel. Technol. 1, 83-90 (2004) doi:10.1089/154065802761001338.
9. www.malvern.com
10. Demarest, S.J., and Frasca. V., Differential Scanning Calorimetry in the Biopharmaceutical Sciences, in: Biophysical Characterization of Proteins in Developing Biopharmaceuticals, D.J. Houde, S.A. Berkowitz (eds.), Elsevier, Amsterdam, Netherlands, p. 287-306 (2015).
11. Remmele, R.L., Microcalorimetric Approaches to Biopharmaceutical Development, in: Analytical Techniques for Biopharmaceutical Development, R. Rodriguez-Diaz, T. Wehr, S. Tuck (eds.), Taylor & Francis, New York, NY, USA, p. 327-381 (2005).
12. Morar-Mitrica, S., Nesta, D., and Crofts, G., BioPharm Asia 2, 46-55 (2013).
13. Kirkitadze, M., Hu, J., Tang, M., and Carpick, B., Pharm. Bioprocess. 2, 491-498 (2014) doi: 10.4155/PBP.14.27.
14. Wen, J., Arthur, K., Chemmalil, L., Muzammil, S., Gabrielson, J., and Jiang, Y., J. Pharm. Sci. 101,955-964 (2012) doi: 10.1002/jps.22820.
15. Gabrielson, J.P., and Weiss, W.F., J. Pharm. Sci. 104, 1240-1245 (2015) doi: 10.1002/jps24393.
16. Cooney, B., Jones, S.D., and Levine, H., BioProcess Int. 14(6), 28-35 (2016). http://www.bioprocessintl.com/analytical/upstream-development/quality-by-design-for-monoclonal-antibodies-part-1-establishing-the-foundations-for-processdevelopment/
17. Cooney, B., Jones, S.D., and Levine, H., BioProcess Int. 14(8), 24-33 (2016). http://www.bioprocessintl.com/2016/quality-design-monoclonal-antibodies-part-2-process-design-space-control-strategies/
18. http://www.who.int/biologicals/biotherapeutics/rDNA_DB_final_19_Nov_2013.pdf
19 http://www.ema.europa.eu/docs/en_GB/document_library/Scientific_guideline/2009/09/WC500003935.pdf
20. http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM496611.pdf
21. Jiang, Y., and Nahri, L.O., J. Am. Pharm. Rev. 9, 34-43 (2006).
22. Arthur, K.K., Dinh, N. , and Gabrielson, J. P., J. Pharm. Sci. 104, 1544-1554 (2015) doi: 10.1002/jps.24313.
23. Shahrokh, Z., Salamat-Miller, N., and Thomas, J.J., Biophysical Analyses Suitable for Chemistry, Manufacturing, and Control Sections of the Biologic License Application (BLA), in: Biophysical Methods for Biotherapeutics: Discovery and Development Applications, T.K. Das (ed.) John Wiley & Sons, Hoboken NJ USA pages 317-353 (2014).
24. Lubiniecki, A., Volkin, D.B., Federici, M, et al., Biologicals 39, 9-22 (2011). doi: 10.1016/j.biologicals.2010.08.004.
25.http://www.fda.gov/Drugs/DevelopmentApprovalProcess/HowDrugsareDevelopedandApproved/ApprovalApplications/TherapeuticBiologicApplications/Biosimilars/
26.http://www.ema.europa.eu/docs/en_GB/document_library/Scientific_guideline/2015/01/WC500180219.pdf
27.http://www.who.int/biologicals/areas/biological_therapeutics/BIOTHERAPEUTICS_FOR_WEB_22APRIL2010.pdf
28 Bas, T.G., and Oliu Castillo, C., Biomed Res Int. 2016, 5910403 (2016). doi: 10.1155/2016/5910403.
29. Tsuruta, L.R., Lopes dos Santos, M., and Moro, A.M., Biotechnol Prog. 31, 1139-1149 (2015). doi: 10.1002/btpr.2066.
30 Kálmán-Szekeres, Z., Olajos, M., and Ganzler, K., J. Pharm. Biomed. Anal. 69, 185-195 (2012). doi: 10.1016/j.jpba.2012.04.037.
31 Berkowitz, S.A., Engen, J.R., Mazzeo, J.R., and Jones, G.B., Nat. Rev. Drug Discov. 11, 527-540 (2012). doi: 10.1038/nrd3746.
32.http://www.fda.gov/downloads/AdvisoryCommittees/CommitteesMeetingMaterials/Drugs/ArthritisAdvisoryCommittee/UCM510493.pdf
33.http://www.fda.gov/downloads/AdvisoryCommittees/CommitteesMeetingMaterials/Drugs/ArthritisAdvisoryCommittee/UCM484860.pdf
34. Jung, S.K., Lee, K. H., Jeon, J. W., et al., MAbs 6, 1163-1177 (2014). doi: 10.4161/mabs.32221.
35.http://www.fda.gov/downloads/AdvisoryCommittees/CommitteesMeetingMaterials/Drugs/ArthritisAdvisoryCommittee/UCM510293.pdf
36. Liu, J., Eris, T., Li, C., Cao, S., and Kuhns, S., BioDrugs 30, 321-338 (2016). doi: 10.1007/s40259-016-0184-3.
37. Sinha-Datta, U., Khan, S., and Wadgaonkar, D., Biosimilars 5, 83-91 (2015) doi: 10.2147/BS.S85537 https://www.dovepress.com/label-free-interaction-analysis-as-a-tool-to-demonstrate-biosimilarity-peer-reviewed-article-BS