A brief technical overview of the system, principals of analysis and applications in protein characterization, process development and interaction analysis.
Differential scanning calorimetry (DSC) is a powerful analytical tool for characterizing the stability of proteins and other biomolecules. DSC directly measures the enthalpy (∆H) and temperature (Tm) of thermally induced structural transitions in solution. This information gives valuable insights into factors that contribute to the conformational stability of macromolecules and complexes such as proteins, DNA and lipid or detergent micelles. This information is used throughout the biotherapeutic industry covering a broad range of applications such as the maximization of drug shelf life, the development of purification strategies and the evaluation and characterization of protein constructs. It is also used in small molecule drug discovery programs, particularly in HTS assay development, hit validation and structural biology groups.
MicroCal VP-Capillary DSC system has an active cell volume of 130 µl allowing for thermodynamic measurements of precious samples (sample concentrations of about 0.2 mg/ml are typically needed).

|
A fully integrated autosampler enables running of up to 50 samples per day and software with automated data analysis features significantly reduces the analysis time. Cell filling, injection, and cleaning functions are fully automated for walk-away operation. For applications where higher throughput and unattended operation and sample handling are not required, MicroCal Manual VP-Capillary DSC is also available with opportunity to upgrade for full automation. MicroCal VP-Capillary DSC system is controlled by MicroCal Control Software and data analysis is performed with MicroCal-enabled Origin® software.
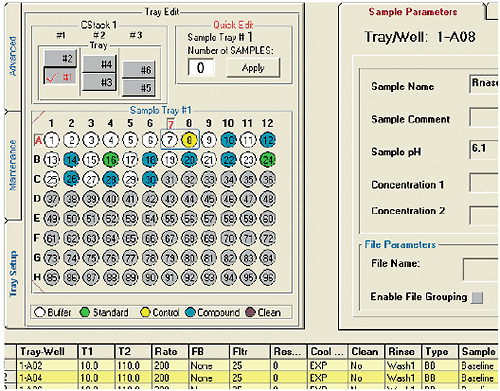
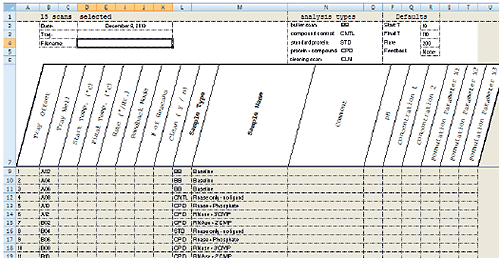
MicroCal VP-Capillary DSC software facilitates experiment design and setup by providing a single, intuitive graphical user interface for selecting and entering all sample and experiment run parameters (Fig 2). For maximum productivity, the software includes a Microsoft® Excel® sample list and experiment template (Fig 3) that allows offline experiment design and setup. A completed template can be saved and reused reducing the time it takes to design and set up future experiments. Additional sample trays can be appended to a running experiment without having to interrupt the run, providing flexibility for multi-user operation. Efficiency in experiment design is further enhanced with intelligent run parameter defaults and optional data fields for completely describing samples.
|
|
The automated data analysis features in MicroCal VP-Capillary DSC software work seamlessly with Origin software to significantly reduce the data analysis bottleneck. Buffer scans and baselines are automatically subtracted and data is automatically normalized to concentration and immediately ready for the user to analyze. The user can also set baseline detection and subtraction limits to select the desired peaks for a particular experiment. All key data parameters, including Tm, ΔTm, ΔH, and Tm onset, are automatically generated and tabulated. All data are easily exported to Microsoft Excel.
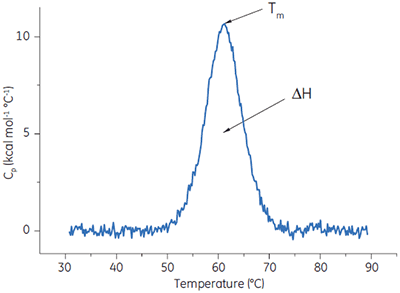
DSC measures the heat absorbed when a protein or other biological macromolecule undergoes a ‘melting’ between a native, biologically active conformation and an unstructured, inactive conformation.
A protein is placed in the calorimetric cell and is heated at scan rates typically between 10°C and 240°C/h. Protein unfolding is an endothermic event and is observed in DSC as a positive displacement in the signal (the heat capacity). The midpoint of this ‘melting’ transition is known as the Tm, and the area under the ‘melting’ curve is the enthalpy, ΔH, of the process (Fig 4). In addition to the classical application of understanding the noncovalent forces responsible for macromolecular assembly and stability, these data can be used to understand the factors that contribute to the folding and stability (shelf life) of the macromolecules.
|
In many cases, decisions can be made on which excipients and buffers will lead to the most favorable conditions for the protein or biotherapeutic simply by observing shifts in the Tm. Shifts to a higher Tm indicate more favorable stabilizing conditions. Strong correlations between Tm data and shelf life are frequently observed. In addition to giving insight into the thermodynamics of protein stability, the area-under-the-curve data (∆H, enthalpy) reflect the extent of folded molecules making DSC ideal for quantitation of the intact protein after storage, purification, or manufacture.
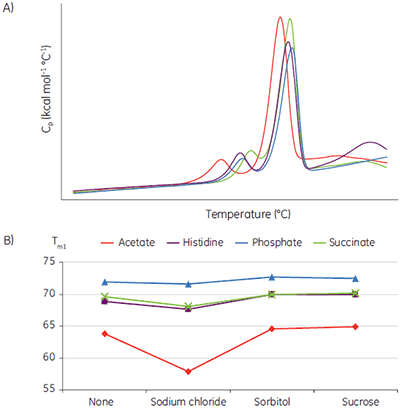
Liquid biotherapeutics are highly desirable because they are easy to administer and less expensive to produce than lyophilized drug product. The challenge is finding conditions where they remain active with no aggregation and no precipitate formation. Tm data has been shown to be an exceptionally good indicator of the relative stability of liquid formulations. Figure 5 shows an example where a MicroCal VP-Capillary DSC system has been used to rapidly screen formulation conditions for a monoclonal antibody (MAb). The stability study was done as part of a preliminary biophysical study and was used to narrow down the solution conditions (pH, buffer and excipient concentration) to be used in the subsequent optimization cycle.
The buffer-excipient combinations containing the sample protein were scanned at 60°C/h, using a protein concentration of 2 mg/ml. The raw data (Fig 5A) was buffer baseline subtracted, the concentrations normalized, and the thermal transitions plotted as a function of buffer types and excipient (Fig 5B).
These data show that acetate buffer is not suitable and a combination with sodium chloride, in particular, should be avoided in further optimization studies.
|
Biotherapeutic downstream production processes must achieve high purity and adequate yield on a large scale and within a given economic framework. Using DSC, the most stabilizing loading and elution conditions can be established during process development. In Figure 6, DSC is used to establish the stability of an antibody in a range of buffers.
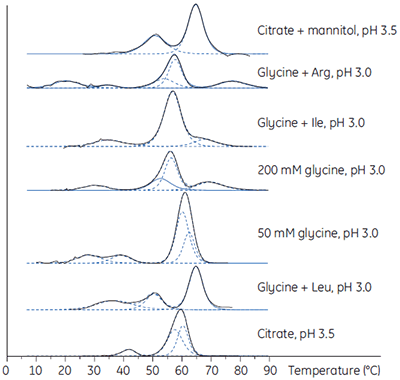
|
The established purification protocol required elution at low pH, a condition in which the antibody was unstable and had a high tendency to aggregate. To minimize the effect, the proteins were loaded and eluted at low concentrations. The DSC study showed that the stability of the antibody was significantly enhanced in the presence of mannitol (see the top DSC profile in Fig 6) and, when included in the elution buffer, resulted in a 7.5-fold increase in the functional capacity of the purification medium and a comparable reduction in the cost to purify the antibody as a result of the better utilization of this purification medium.
When a drug candidate binds to the target protein, the Tm of the drug-target complex is shifted relative to that of the target molecule alone. The binding constant can be calculated from the Tm shift at a single ligand concentration (1). The expressions describing this relationship are in the software provided with MicroCal VP-Capillary DSC instrument. Ribonuclease A (RNAse A, 0.03 mM in 50 mM potassium acetate, pH 5.5) was studied with MicroCal VP-Capillary DSC in the presence of 0, 0.5, and 1 mM of 2’CMP, and data from a series of 96 measurements performed at a scan rate of 240°C/h taken over 2 days (approx. 50/day) are shown in Figure 7.
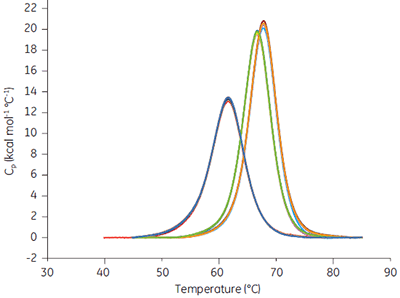
|
The scans were automatically analyzed to generate Tm and ∆H values. Tm was 61.62 ± 0.06°C in the absence of 2’CMP, 66.7 ± 0.03°C in the presence of 0.5 mM 2’CMP, and 67.77 ± 0.03°C in the presence of 1 mM 2’CMP. The binding constant determined at both concentrations was approx. 1 µM, at 25°C, under both conditions and compares favorably with direct isothermal titration calorimetry (ITC) measurements.
The relationship between affinity and Tm shift means that DSC can be used to rank order the affinities of related compounds. Tm data for RNase (0.09 mM in 50 mM acetate buffer, pH 5.5) obtained with MicroCal VP-Capillary DSC at 200°C/h in the presence of a number of ligands at a concentration of 10 mM are shown in Table 1 together with Kd as determined by ITC at 25°C under the same buffer conditions.
| Tm (°C) | Kd (µM) at 25°C | |
|---|---|---|
| No ligand | 61.08 | - |
| Phosphate | 62.95 | 122 |
| 3’UMP | 67.47 | 26 |
| Pyrophosphate | 68.24 | 12 |
| 2’CMP | 70.71 | 1.7 |
The rank order of Tm data generated by DSC is the same as the affinity data determined directly by ITC, demonstrating the utility of DSC for comparing the binding of related ligands. It should be noted that certain assumptions may need to be made for the quantitative determination of Kd using this method, and that these values should be considered as approximate values in all but the most favorable instances.
The magnitude of the Tm shift is related to both the concentration and affinity of the added ligand. A weak binder will typically induce a smaller Tm shift than a tight binder at the same concentration. This has practical consequences for those involved in fragment-based or more traditional high-throughput screening campaigns in small molecule drug discovery. To screen for the weak (approx. 1 mM) binders typical for fragment-based programs, the DSC measurements would require ligand concentrations close to 1 mM to cause a 1°C shift. In more traditional hit-validation campaigns, ligand concentrations close to 50 µM could be used. Compounds with affinities of 5 µM or lower would induce Tm shifts of 2°C or more during these circumstances.
1. Brandts, J.F. and Lin, L.N. Study of strong to ultratight protein interactions using differential scanning calorimetry. Biochemistry 29, 6927-6940 (1990).
Data on stability screening and Figures 5A and 5B were kindly provided by Alex Tracey, Novartis (formerly with KBI Biopharma). Data on optimization of purification conditions and Figure 6 were kindly provided by Prathima Acharya, Biosonata Consulting (formerly with Diosynth Biotecnhologies).
| Cell volume | 130 μl |
| Sample volume | 370 μl |
| Typical sample concentration | 0.1 to 2 mg/ml |
| Response time | 5 s* |
| Noise | 0.05 μCal/°C† |
| Baseline repeatability | 1.5 μCal/°C‡ |
| Multiple feedback modes | Yes (passive, high gain, low gain) |
| Temperature range | -10°C to 130°C§ |
| Maximum scan rate | 240°C/h |
| Cell design | Tantalum, capillary type, fixed-in-place, non-removable |
| Cell to cell heat compensation | Power feedback |
| Throughput | Up to 50 samples/24 h |
| Sample capacity | Six 96-well plates |
| Self-contained pressuring system | 0 to 45 psi |
| Automation upgrade available | Yes |
| Weight | 25 kg |
| Dimensions (W × H × D) | 101 × 70 × 68 cm |
* High feedback.
† Using upscan mode at 200°C/h with 10 s filter and over a temperature range of 10°C to 110°C, in passive mode.
‡ Average standard deviation between successive upscans at 90°C/h with 30 s data point filter over the temperature range 5°C to 110°C.