TDA with UV-detection is used to follow self-association of protein-based systems, and critically is able resolve the smaller monomeric species since measurements are not adversely affected by the presence of a small amount of oligomer in mixtures.
Taylor Dispersion Analysis (TDA) is an orthogonal technique for sizing and stability studies of biomolecules in solution. Studies have shown that hydrodynamic radius (Rh) measurements from TDA on monodisperse unstressed protein-based systems e.g. monoclonal antibodies (mAbs), are comparable to data obtained using the well-established and complementary technique of Dynamic Light Scattering (DLS)1,2.
Whilst both TDA and DLS can pick up changes in self-association and oligomeric state between samples, the two techniques respond differently to the presence of mixtures.
DLS is very sensitive to the presence of large species, but this measurement response can mask information about the smaller species in the sample. In contrast, TDA measurements in mixtures are able to resolve the smaller species and are not adversely affected by the presence of a small amount of a larger species.
As an important biotherapeutic used in the treatment of Diabetes Mellitus, Insulin provides an interesting case study with which to review the use of TDA as a technique for characterizing the hydrodynamic radius (Rh) and stability of protein-based biopharmaceuticals in solution, and to highlight the differences in data obtained for oligomeric mixtures from DLS measurements.
At neutral pH, Insulin oligomers have been shown to dissociate at low protein concentrations, a feature that is key to their function within the body3. Hexameric Insulin is the more stable form used to store the protein within cells, whereas monomeric Insulin is the active form that binds to Insulin receptors. Identifying the oligomeric state of Insulin in a specific formulation is therefore important in predicting the stability of the formulation. Hexamers are formed through the self-association of Insulin monomers into dimers, which combine to form hexamers. Changes in
Insulin’s oligomeric state can be observed as a change in size, where smaller sizes correspond to lower order oligomers.
TDA is an absolute method for determining hydrodynamic size which is achieved by monitoring the dispersion of a nanoliter volume sample pulse as it flows through a microcapillary. The sample pulse broadens due to a combination of dispersion and diffusion, to an extent which is a function of the hydrodynamic size of the solute molecules. The time-evolved concentration profile (or Taylorgram), measured by UV absorption at a specific wavelength, is monitored at two fixed points along the microcapillary. Analysis of the Taylorgrams enables the molecular diffusion coefficient and hence hydrodynamic radius of solute molecules to be determined.
The following work details measurements to monitor self-association and oligomeric state of Insulin in solution with decreasing concentration at neutral pH.
Example Taylorgrams for Insulin hexameric and monomeric samples are given in Figure 1, showing changes in peak width with hydrodynamic radius - information which is used to determine the hydrodynamic radius of the molecule. Molecules with a larger hydrodynamic radius have wider peaks, whilst peak widths for molecules with a smaller hydrodynamic radius are narrower4.
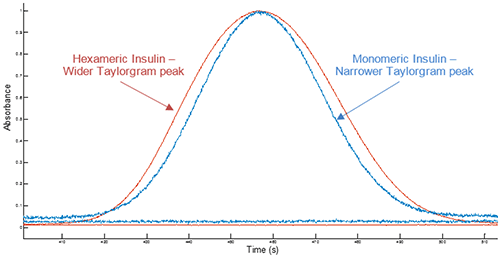
Figure 1 Overlay of example Taylorgrams (UV absorbance vs time) from Viscosizer measurements for samples of hexameric Insulin and monomeric Insulin (note Absorbance normalized to 1)
Figure 2 shows the hydrodynamic radius (Rh) in PBS for a series of Insulin concentrations as measured by both Taylor Dispersion Analysis (TDA) and Dynamic Light Scattering (DLS).
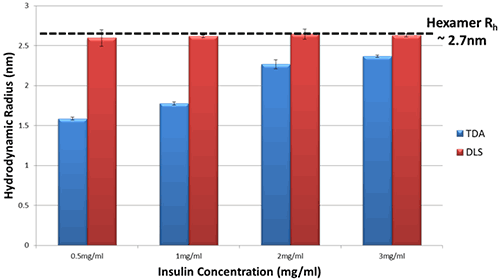
Figure 2 Hydrodynamic Radius measurements for a series of different Insulin concentrations in PBS buffer at 20°C as determined by TDA and DLS
Changes in hydrodynamic radius can be used to indicate a change in the self-association state of a molecule.
The DLS data show essentially no change in hydrodynamic radius of Insulin in PBS, within the limits of experimental uncertainty.
In comparison, the TDA data show a clear difference in hydrodynamic radius of Insulin, and a distinct trend with concentration. As the concentration of Insulin decreases, the observed size decreases markedly thereby implying a decrease in oligomeric state.
To corroborate a size reduction with concentration for Insulin in PBS, the samples were also assessed using Size Exclusion Chromatography (SEC). SEC data shows an increase in retention volume as the concentration of Insulin decreases and therefore a decrease in Insulin size.
This difference in data arises because DLS is an intensity-weighted technique and therefore sizes observed by DLS are biased towards the size of the largest oligomer present within the sample; in this case, the Insulin hexamer.
By comparison, TDA with UV detection is a mass-weighted technique and therefore offers measurement benefits when identifying small changes to a molecule of interest.
As highlighted in the data for Insulin solutions in Figure 2, utilizing comparable data from TDA can offer new insight into self-association state of molecules, as the measurements are not adversely affected by the presence of a small amount of larger species or aggregates in solution.
Using Insulin as a case study, this Application Note demonstrates how TDA can be used to monitor size changes induced by changes in oligomeric state of a protein with a change in concentration. It also shows how TDA can be used to provide complementary size information to existing techniques such as DLS, with the advantages of its inherent capability to track changes in oligomeric state without adverse bias from larger species or aggregates in solution (due to mass-weighted detection).