The Zetasizer μV is a dynamic and static light scattering (DLS & SLS) system capable of making measurements of molecular size and molecular weight in a batch cuvette and also when connected to a size-exclusion chromatography (SEC) system in conjunction with an OmniFACE connection box (figure 1).

|
The Zetasizer μV has a 90° detection angle with excellent signal-to-noise and a variable power laser and attenuator to help it to achieve a wide dynamic range with respect to the light scattering count rate. Additionally, the light scattering volume, which is the intersection of the illuminating laser and detector field, is very small. This allows very small volumes of sample to be measured. In a batch cuvette, measurements can be made in as little as 2 μl while the flow cell used with the system is just 8 μl. Switching between batch and chromatography modes takes only seconds for the removal of the batch cuvette and insertion of the flow cuvette or vice versa.
The application arena for the Zetasizer μV is primarily the characterization of proteins and the design is such that it can be interfaced with almost any third-party SEC system such as the commonly used GE AKTA. This application note describes a selection of measurements made on a GE AKTA basic system. Since the unit only contains a UV detector for measuring sample concentration, this can be used for the molecular weight calculations instead of the more commonly used refractive index detector, provided that the molar extinction coefficient of the sample protein is known. If this is not known, then reliable estimates can be made based on the amino acid sequence of the sample.
Measurements were made using a GE AKTA basic system with a GE Superdex 200 column, a typical SEC setup used in many biochemical laboratories. The mobile phase used was phosphate buffered saline.
The Zetasizer μV was plumbed in line after the UV cell in the AKTA. The analogue signal from the UV detector was connected to the OmniSEC software via the OmniFACE interface board.
Bovine serum albumin is commonly used as a standard protein for system calibration. It has a well established molecular weight and molar extinction coefficient meaning both the UV detector and the Zetasizer μV can be calibrated using this one sample which is in plentiful supply and well characterized.
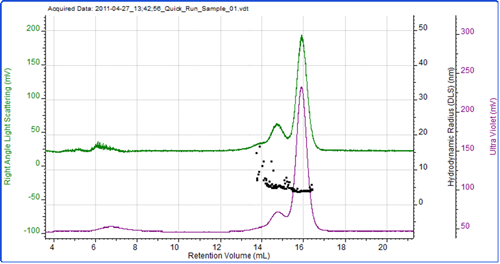
Figure 2 shows a chromatogram of BSA with the UV trace in purple coming from the UV detector in the AKTA and the green trace showing the light scattering intensity as measured by the Zetasizer μV. As is common with this protein, the sample has eluted as a number of peaks which represent the monomer and various oligomers. As the sample was eluting, the hydrodynamic size was measured using the correlation functions collected during the run at 3 second intervals. The individual size measurements can be seen plotted on the chromatogram.
|
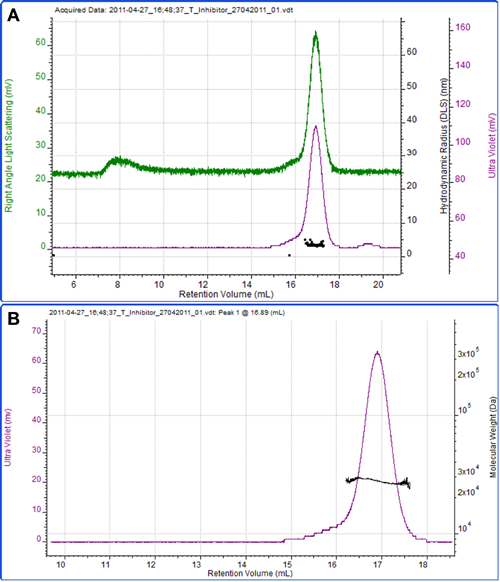
Figure 3 shows a measurement of another protein, soy bean trypsin inhibitor. A theoretical extinction coefficient of 0.4 ml mg-1 cm-1 at 280nm was used to measure the sample concentration. A dn/dc of 0.185 ml/g, which is commonly accepted to be constant between many proteins, was also used. The calculated molecular weight is 27.5 kDa, which is in excellent agreement with the theoretical molecular weight of 28 kDa. This clearly identifies this protein as the monomer. The hydrodynamic radius was also measured by DLS and the mean size reported as 3.3 nm, which is reasonable for a protein of this molecular weight.
|
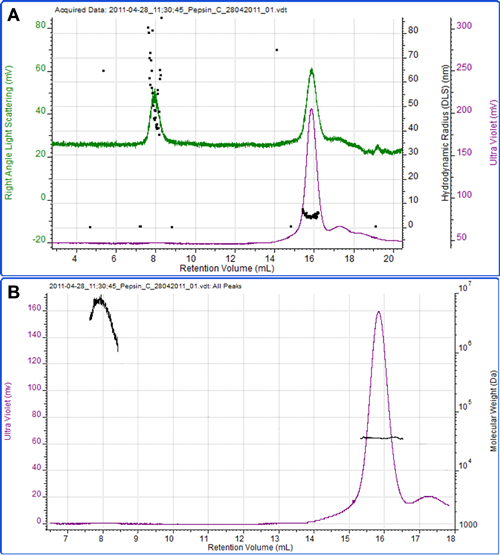
Figure 4 shows a final example of the protein pepsin. Using an extinction coefficient of 1.47 ml mg-1 cm-1 at 280nm, the concentration was measured and the molecular weight calculated as 35.2 kDa which is in excellent agreement to the known molecular weight of the protein of 35 kDa. The radius measured by DLS was 4.6 nm.
|
A second peak eluted at 8 ml, which is most notable on the light scattering trace. The high size and molecular weight and the polydispersity of this peak clearly identify it as some aggregated material within the sample.
The Zetasizer μV is capable of making DLS measurements of hydrodynamic radius and SLS measurements of molecular weight in both cuvette and chromatography setups.
This application note has demonstrated that it is possible to make these measurements when the system is connected to a third-party SEC system such as the GE AKTA.
ÄKTA is a trademark of GE. Healthcare Bio-Sciences AB.