This application note presents the characterization of a protein at high concentration. When the average interparticle distance is smaller than the screening length, unexpected size results can appear due to electro-static protein interactions.
TAKA Amylase is an enzyme involved in the digestion of amylase. The molecular weight of this protein is 53 kDa, and its structure is shown in figure 1.
|
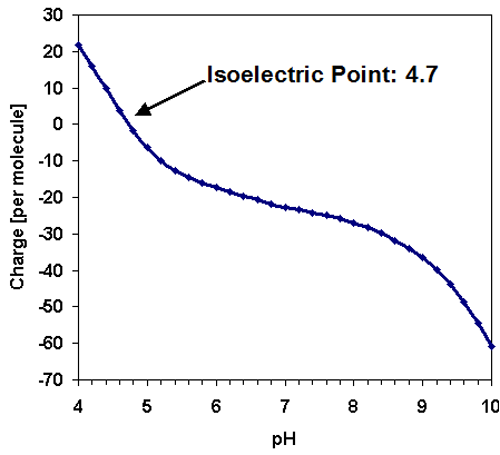
Like the majority of proteins, the amylase molecule is expected to be negatively charged at neutral pH conditions. The predicted isoelectric point [1] of TAKA Amylase is in the acidic region at 4.7 and the charge per molecule is displayed as a function of pH in figure 2.
|
Concentrated samples of TAKA Amylase were prepared at 20 mg/mL and 100 mg/mL in water. It is normally not advisable to dissolve proteins in pure water, however, at such high concentrations enough counter ions are present in the molecule to prevent aggregation. Solutions are clear and stable. An additional sample at 20mg/mL was prepared in 100mM Tris buffer at pH 8 to compare with the water only sample. Measurements were performed using a Zetasizer Nano without filtering the sample.
All samples were clear. The protein was at such high concentration that the scattering intensity detected was adjusted automatically using the auto attenuator in the instrument.
Dynamic light scattering (DLS) was used to analyze the fluctuations in the scattered light leading to the determination of the protein diffusion coefficient. Applying knowledge of the viscosity of the diffusing medium (water in this study), a hydrodynamic size of the molecule was obtained. This is the size of a sphere which diffuses with the same diffusion coefficient as the protein.
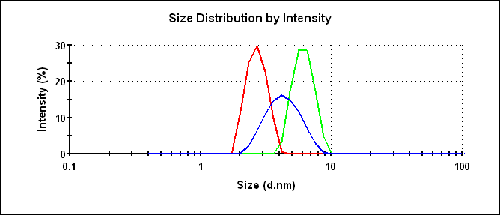
The size results for the three different samples are shown in figure 3 and listed in table 1. The buffer consisted of 100mM Tris at pH 8.0. The protein at 20 g/L provides a reasonable prediction of the molecular weight under dilute conditions.
|
| Concentration (g/L) | Solvent | z-Average Diameter (nm) | Protein Peak (nm) |
|---|---|---|---|
| 20 | Buffer | 6.1 | 6.2 |
| 20 | Water | 2.6 | 2.7 |
| 100 | Water | 5.1 | 4.4 |
The expected size for the molecule is 6.5nm assuming an empirical model for average globular proteins. Only the measurements of TAKA Amylase in the buffer are close to the expected value.
The intensity size distributions in figure 3 clearly show that the two preparations in water produce a smaller measured size than in the buffer.
The absolute molecular weight is determined by detecting the scattering signal from known concentrations of the sample and using a Debye plot to analyze the data. This measurement provides the weight-average molecular weight of all species in solution.
The refractive index increment dn/dc of many proteins is approximately 0.185 mL/g. Using the derived intensity values from the two concentrations in water the estimated average molecular weight is 14 kDa which is obviously much lower than the known molecular weight of 53kDa. Although SLS should ideally be performed with more concentrations, this result again underscores the strong effects of the solvent on the light scattering analysis of biomolecules.
This example of TAKA Amylase shows that the choice of solvent is very important when performing light scattering experiments, as the protein exhibits very unusual properties when dissolved in deionized water. For practical applications, protein samples should best be prepared in buffers. The lack of ions in deionized water can lead to size and molecular weight results that may be difficult to interpret.
The Zetasizer Nano system from Malvern Instruments is the first commercial instrument to include the hardware and software for combined dynamic, static, and electrophoretic light scattering measurements, providing the researcher with a wide range of sample properties, including the size, molecular weight, and zeta potential. The system was specifically designed to meet the low concentration and sample volume requirements typically associated with pharmaceutical and biomolecular applications.
References
[1] http://www.scripps.edu/~cdputnam/protcalc.html