Size-exclusion chromatography (SEC) is an essential tool for the characterization of proteins for academic research or in the development of biopharmaceutical drugs.
While single-detector SEC systems are widely used in the characterization of proteins, particularly for the detection of protein aggregates, multi-detector systems are a more advanced option which brings numerous advantages.
Single-detector systems might typically include only a ultraviolet (UV) detector, and would be used to study proteins and to identify the presence of any aggregates. They are sometimes also used to make estimates of molecular weight by comparing a protein’s elution volume against that of standards of known molecular weights.
Single-detector conventional systems are widely used and are popular for a number of reasons including their repeatability and reproducibility. That repeatability though is due to the fact that many underlying changes in protein molecular weight and structure are masked by the technique’s limitations.
In recent years, however, technology has advanced and with additional detectors comes a wealth of additional information. Light scattering detectors, whether SEC-MALS (multi-angle light scattering), RALS (right-angle light scattering), or LALS (low-angle light scattering), measure absolute molecular weight with excellent accuracy. This allows oligomers and larger aggregates to be easily differentiated and quantified. Conjugate analyses allow the characterization of PEGylated proteins, antibody-drug-conjugates (ADCs) and membrane proteins.
A viscometer is occasionally used to study large changes in protein conformation.
With these capabilities in mind, this white paper lists the top ten reasons why your laboratory would benefit from an advanced multi-detector SEC system.

|
|
The greatest limitation of conventional SEC measurements is the way in which they calculate molecular weight. Since molecular weight is calculated from retention volume (which is directly related to the sample’s size) and a comparison with molecular weight standards (such as a series of globular proteins), the calculated results are only accurate if the protein’s structure is globular and happens to fit on the calibration curve. Conventional calibration is blind to countless possibilities of a protein’s structure and the effects this have on its elution time.
With so many proteins under study, the molecular weight of only very few can be accurately measured with such a technique. It is far more common for researchers to simply assume that the primary peak in a separation is the monomer and that anything else present is some form of aggregate. While this may be sufficient for a few applications, it is an extremely limited amount of information. The addition of a light scattering detector means that from now on, all of your molecular weight measurements will be accurate, irrespective of the protein being measured. Absolute molecular weight means a better understanding of your protein and the types of aggregates formed, providing better control over its behavior and stability. Ultimately, this ensures a better quality product. In research, absolute molecular weight means better knowledge about a newly purified or recombinant protein, and greater accuracy in data used for publication purposes.
One of the key reasons for measuring the absolute molecular weight of a protein is to reliably assess its oligomeric state. Many proteins come together in multimers of two or more sub-units to control activity and/or stability. For instance, insulin, while active as a monomer, forms an inactive hexamer for stability during production and storage.
In a biopharmaceutical, the difference between oligomeric states can mean the difference between stability and rapid aggregation, or between activity and inactivity. In research, it can mean the difference between a functioning experiment and useful data, and a significant amount of wasted time and effort.
Multi-detector SEC and light scattering, in particular, allow accurate measurement of protein oligomeric state. This allows you to better understand the differences between stability and aggregate formation, or activity and inactivity.
In the example in Figure 2, a chromatogram of carbonic anhydrase is shown. This sample consists of four populations - peaks 2, 3, and 4 can easily be identified as the trimer, dimer, and monomer respectively, thanks to the accuracy of the measured molecular weights (shown in Table 1). The stability of the measured molecular weight across the peaks clearly shows that they are distinct populations. The molecular weight of peak 1 is higher and more varied, which identifies it as larger agglomerates. The fraction of each peak in the total sample can also be measured. In this way, the sample is far more accurately and clearly characterized than it could have been with a single-detector system.
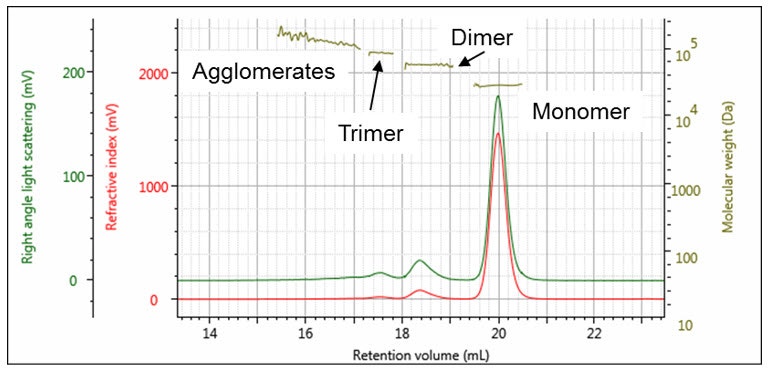
Figure 2: Chromatogram of carbonic anhydrase with associated measured molecular weights
| Peak | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| RV (mL) | 17.04 | 17.54 | 18.37 | 20.01 |
| Mw (g/mol) | 125,800 | 87,640 | 58,750 | 29,200 |
| Mw/Mn | 1.019 | 1.000 | 1.000 | 1.000 |
| Frac. of sample (%) | 0.883 | 1.311 | 6.439 | 91.37 |
| RI peak (mV.mL) | 5.625 | 8.321 | 40.89 | 580.9 |
| (RALS peak (mV.mL) | 2.889 | 2.982 | 9.825 | 69.39
|
There is no such thing as a typical protein aggregate. They come in all shapes and sizes, structures and conformations. For instance, aggregates of amyloid proteins are known to form fibers that are present in diseases such as Alzheimer’s. Many other proteins form aggregates which are dense and compact particulates. Proteins like β-lactoglobulin will form aggregates of different structures depending on conditions.
Oligomers themselves are often included in the wider description of ‘aggregates’. Oligomers are typically differentiated from other aggregates by their stable molecular weight and activity and are easily identified by their molecular weight which is always a multiple of the monomer’s.
Larger and more disordered aggregates of different structures will have different properties. Some, for example amyloid proteins, are involved in specific disease states. Others may have varying immunogenic effects when injected as part of a biotherapeutic.
Multi-detector SEC can go beyond simple detection of unidentified aggregates within a sample. It can measure their molecular weight and polydispersity (whether they are one molecular weight or cover a range of weights). It can tell us if those aggregates are globular, particulate and small, or large, extended and fibrous. Such information can help identify mechanisms of aggregation as well as pinpoint where in production the aggregation might be occurring.
In Figure 3, the aggregates in an IgG sample are clearly separated from the monomer for molecular weight measurement. Those eluting at 10 – 11 minutes are barely visible on the RI detector but clearly present on the light scattering signal demonstrating its sensitivity to these aggregates.
Our customers are using multi-detector systems to characterize samples exactly like this in biopharmaceutical manufacturing.
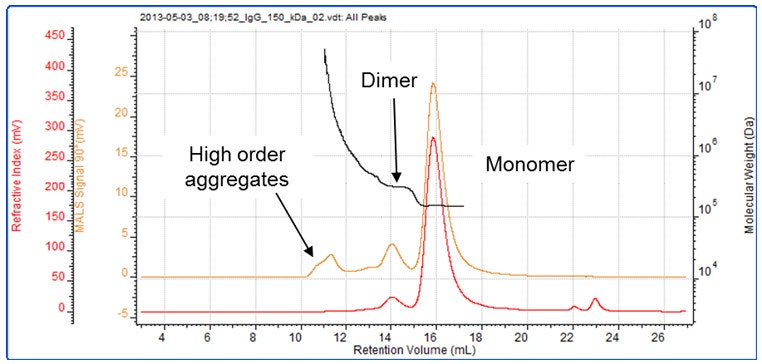
Figure 3: A sample of IgG contains, monomer, dimer and higher order aggregates
One key advantage of a single-detector conventional calibration system is its repeatability. However, that repeatability comes from the fact that it only provides limited information. There may well be underlying differences between your samples which are not visible when using UV-SEC alone. For instance, what appears to be a simple aggregate peak could be an oligomer or a larger, more disordered aggregate. In an assay, or when administered to a patient, the different responses to these two types of aggregate could be significant. Oligomers may have more or less activity than the monomer alone, while larger, disordered aggregates will reduce efficacy (through reduction of active molecules) and can have immunogenic side-effects. It may be that one of these types of aggregates is acceptable while the other is not, meaning that some batches may be rejected despite being functional or, worse, others may be accepted when in fact they will cause problems.
As an example, the chromatogram of β-amylase in Figure 4 appears to show a protein with some aggregates. However, upon closer examination with a multi-detection SEC system, peak 2 is revealed to be a protein of similar molecular weight. Its elution volume is not the same as the β-amylase because it has a dramatically different structure, as evidenced by the measured intrinsic viscosity and hydrodynamic radius. Comparing the UV and RI signals reveals it to be a contaminating protein, as opposed to a structural variant of the β-amylase. 1
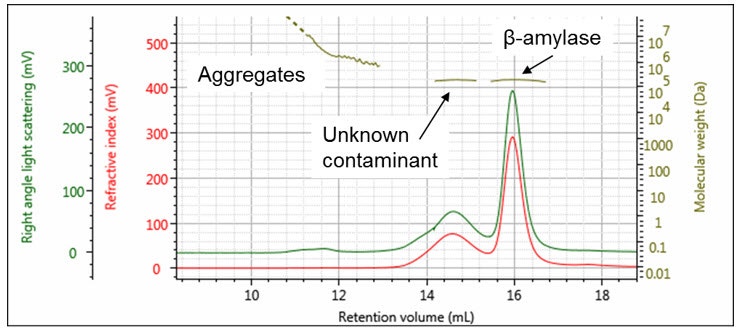
Figure 4: Chromatogram of β-amylase with associated measured molecular weights. Intrinsic viscosity and hydrodynamic radius.
| Peak | 1 | 2 | 3 |
|---|---|---|---|
| RV (mL) | 12.93 | 14.60 | 15.97 |
| Mw (g/mol) | 9,543,000 | 208,500 | 212,800 |
| Mw/Mn | 6.138 | 1.001 | 1.001 |
| IVw (dL/g) | N/C | 0.1728 | 0.06318 |
| Rh(η)w (nm) | N/C | 8.283 | 6.000 |
| Frac. of sample (%) | 0.3573 | 28.19 | 71.45 |
| RI peak (mV.mL) | 0.7691 | 62.07 | 157.3 |
| RALS peak (mV.mL) | 7.389 | 52.49 | 136.8 |
Assumptions about aggregation based on single-detector measurements can lead to incorrect conclusions about a particular protein sample. A multi-detector system clarifies these differences to help you fully characterize your protein sample. This means a better understanding of its activity and stability, higher-performing drugs and more predictable assays.
Protein conjugates are complex. Conjugates vary both in terms of the number of protein molecules and also the number of conjugate molecules that make up the complex. This may be the number of conjugated drug or PEG molecules in ADCs or in PEGylated proteins, which will determine efficacy, half-life, and transportability, or it may be the amount of detergent attached to a membrane protein after purification, which will affect its ability to crystallize.
A multi-detector system, especially one which contains two concentration detectors, can use the different responses of each detector to each component of the conjugate to deconvolute the concentration of each component. This allows an accurate measurement of the complex’s molecular weight and ultimately, the number of conjugate molecules attached.
This work can help tailor an ADC so that it will be stable, active, and have the desired half-life. Alternatively, our customers are using these systems with synchrotrons to characterize membrane proteins. With it, they are guiding purification of membrane proteins to accelerate research into their structure and minimize cost by reducing the number of assays and the amount of protein required for successful crystallization.
In the example below, another one of our customers is using conjugate composition analysis to study their PEGylated proteins, for conjugate, free PEG and free protein content.2

Figure 5: Multi-detection can identify and characterize free protein, free PEG and protein conjugate in a mixed sample
Intrinsic viscosity is a measure of a molecule’s structure, conformation and density. In the case of proteins, this is linked directly to their globularity. Standard proteins such as those found in standard molecular weight kits are globular and have a low intrinsic viscosity (high density). However, others such as antibodies or intrinsically disordered proteins have non-globular, or extended structures (lower density).
By characterizing the intrinsic viscosity of a protein, it is possible to gather information on the protein’s conformation and density. Large conformational changes which take place as a result of environmental changes or the binding of other molecules can also be monitored to detect the folding of an intrinsically disordered protein. With protein activity so dependent on correct folding and structure, intrinsic viscosity is a powerful tool for characterization of this behavior.
Ultimately, information on intrinsic viscosity can be combined with other measurements (for instance, from Dynamic Light Scattering (DLS) or Analytical Ultracentrifugation (AUC)) to study protein shape and hydration state. With all this information, our customers are making excellent measurements of protein structural and hydration changes in response to molecular interactions.3
This provides a powerful tool for research when studying a protein’s conformation in solution.
Multi-detector SEC could be one of the most data-rich techniques in your lab. It allows for complete characterization of protein samples, which is crucial to understand the activity of proteins in assays and their efficacy as biopharmaceutical drugs. Table 3 shows the parameters measured by multi-detector SEC that are not measured by single-detector measurements.
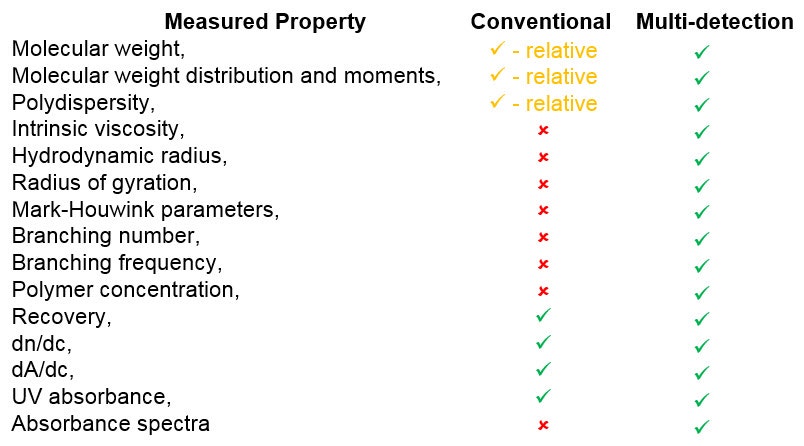
Table 3: Molecular parameters measured by multi-detection vs those measured by single-detector measurements.
With such a large amount of information provided by a single system, advanced SEC is an invaluable technique for your current and future characterization requirements, wherever they may take you.
Many biopharmaceutical labs are using multi-detection-SEC as a tool for diagnosing production issues. While the QC lab uses a simple UV-based single detector system, rejected batches are forwarded to a diagnostic team for further characterization. By studying the molecular weight, size and structure of the proteins and their aggregates, they are able to diagnose the likely source of the aggregation in the production chain.
This helps to identify and resolve quality issues more rapidly. It can be used to help build quality and robustness into a production pipeline to support quality-by-design (QbD) initiatives, saving time and cost in resolving problems and reducing the likelihood of their reoccurrence in future.
Regulation in the biopharmaceutical world is understandably stringent, and new biotherapeutics entering the market must undergo extremely rigorous characterization and production testing.
Where biologic molecules are used as therapeutics, the FDA is increasingly frequently demanding light scattering analysis to accompany UV chromatograms. This provides confidence that the source and properties of any aggregates in the sample are well-understood and that their risk to the patient is minimized. Our customers have been asked directly by the FDA for additional measurements, for example from a light scattering detector, with their single-detector measurements.
Proactive analysis of biologics during their development using light scattering will help reduce the time and cost of bringing a drug to market by demonstrating a keen understanding of your product and its behavior.
Thousands of papers are published every year citing light scattering and chromatography, or SEC-MALS, or multi-detector GPC, or multi-detector SEC. These are published by researchers who are already using these technologies to maximize their productivity. In doing so, they are publishing higher impact research more frequently and they are staying at the forefront of protein characterization research.
In industry, companies are increasingly using multi-detection within R&D to help study protein stability and activity. In manufacturing, these technologies are being used to monitor manufacturing processes or to quickly diagnose and resolve quality issues raised by QC.
In a world of tightening margins and tough competition to sell and to publish, better characterization of proteins and better control of their production means a better and more reliable product. A better product means fewer quality issues, less waste, and better control of manufacturing costs.
If you are characterizing proteins from different drug formulations, quality checking and diagnosing manufacturing issues, multi-detection is a tool to help you work faster and more effectively. This ensures that biopharmaceuticals are stable for longer, have in-built quality, and production issues are resolved quickly.
If you work in research, demands on publishing time and quality mean you don’t have time to repeat purifications and assays because of poorly-understood failures and variable protein activity. A multi-detection experiment means increasingly accurate results faster. This means more powerful articles in higher impact journals and a stronger case to justify the next grant application.
Whatever your industry, the best way to stay ahead of your competition is to have the best information from the most advanced system. Investing in multi-detection is the best way to accomplish this.
Whether you are manufacturing biopharmaceuticals in industry, using them in formulations, or working in research purifying and characterizing novel proteins, multi-detection provides a huge range of advantages over its single-detector predecessor. The sum of all of these advantages is a better understanding of the nature of the protein you are characterizing. With that comes better product quality and control, more valuable and more reliable products and happier customers. In research, it means accelerated development, increasing publication rate, quality and impact.
Malvern Panalytical application note: Compositional insights for protein mixtures: a case study of a semi-pure β-amylase extract.
Malvern Panalytical application note: PEGylated protein: Optimizing conjugation reactions using triple detection SEC.
Calcium-induced folding of intrinsically disordered repeat-in-toxin (RTX) motifs via changes of protein charges and oligomerization states. Sotomayor-Perez et al. 2011.The Journal of Biological Chemistry Vol.286, pp. 16997.