Choosing the best antibody to progress in your biologic pipeline.
Temperature-induced unfolding of three humanized IgG1 monoclonal antibodies and their Fab and Fc fragments was monitored by differential scanning calorimetry at neutral pH. With some exceptions, the thermogram of the intact antibody presents two peaks and the transition with the larger experimental enthalpy contains the contribution from the Fab fragments. Although the measured enthalpy was similar for all three Fab fragments studied, the apparent melting temperatures were found to vary significantly, even for Fab fragments originating from the same human germline. Therefore, we propose to use the measured enthalpy of unfolding as the key parameter to recognize the unfolding events in the melting profile of an intact IgG1 antibody. If the variable domain sequences, resulting from Complementarity Determining Regions (CDRs) grafting and humanization, destabilize the Fab fragment with respect to the CH3 domain, the first transition represents the unfolding of the Fab fragment and the CH2 domain, while the second transition represents CH3 domain unfolding. Otherwise, the first transition represents CH2 domain unfolding, and the second transition represents the unfolding of the Fab fragment and the CH3 domain. In some cases, the DSC profile may present three transitions, with the Fab unfolding occurring at distinct temperatures compared to the melting of the CH2 and CH3 domains. If the DSC profile of a humanized IgG1 monoclonal antibody cannot be described by the model above, the result may be an indication of significant structural heterogeneity and/or of disruption of the Fab cooperative unfolding. Low stability or heterogeneity of the Fab fragment may prove problematic for long-term storage or consistency of production. Therefore, understanding the features of a DSC profile is important for clone selection and process maturation in the early stages of development of therapeutic monoclonal antibodies.
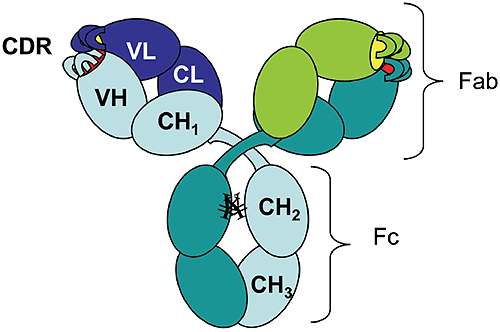
The rapid growth of the therapeutic antibody market has increased the interest in modeling of the antibody structure and in understanding the factors that affect the function and the stability of antibodies. As of 2006 there were 18 monoclonal antibodies approved for therapeutic use in US, and 14 of them were molecules of the IgG isotype.1 Among these, 50% are humanized IgG1 anti-bodies. The antibody molecule (Figure 1) is formed by two identical heavy chains (~450 residues each) and two identical light chains (~220 residues each).
|
The chains fold into domains, of about 110 residues, that have a characteristic beta-sheet architecture known as the "immunoglobulin fold." There are two immunoglobulin domains in the light chain, VL and CL, and four immunoglobulin domains in the heavy chain, VH, CH1, CH2, and CH3 ("V" and "C" stand for "variable" and "constant," respectively). Digestion of human IgG1 antibody by papain results in two types of fragments of similar size: the Fc fragment (composed of two CH2 and two CH3 domains) and the Fab fragment (composed of CH1, CL, VH, and VL).
Mouse-derived antibodies are often subject to immune recognition when administered to humans and their effector functions governed by species-specific Fc fragments are diminished. This has been overcome by "humanization" of murine antibodies, which involves the transfer of complementarity determining regions (CDRs) from the murine antibody along with some adjacent framework regions into the variable domains of the heavy and light chains of a human antibody.2 To identify the key residues to be transferred with CDRs, computer models are used to generate the 3-D structure of the variable domains that can incorporate the CDRs, while preserving the binding specificity and affinity for the desired target.3 There is a limited number of existing human variable domain frameworks (coded in the germline),4 and the selection of the framework is done based on the sequence homology with the murine antibody.2
Temperature-induced unfolding of monoclonal antibodies measured by differential scanning calorimetry (DSC) has become an indispensable tool used for monitoring production consistency, clone selection, protein structure characterization, and may potentially have a value for formulation development. Consequently, correct interpretation of the observed transitions is essential. Temperature or pH-induced unfolding of intact antibodies has been studied, however, mostly with antibodies of non-human origin, such as murine5-8 or rabbit9,10 immunoglobulins. Temperature-induced unfolding of human IgG1 and its Fab and Fc fragments was studied with the myeloma IgG1 Van by DSC in the pH range 3.5-5.5 and compared to the thermograms obtained from other human isotypes (IgG2, IgG3, and IgG4).11
The unfolding of the human myeloma IgG1 at pH 5.5 presents two transitions with the melting temperatures (Tm) around 70°C and 82°C, and with the amplitude of the first peak much larger than that of the second peak. Studies on the Fab and Fc fragments of the same antibody reported a melting temperature of 70°C for the Fab fragment11 and melting temperatures of 66°C and 82°C for the Fc fragment.12 Based on these results one may infer that the first unfolding event in the intact human IgG1 antibody is associated with the melting of the CH2 domain in the Fc fragment and the melting of the Fab fragment, while the second transition represents mainly the unfolding of the CH3 domain. Analogous studies using murine antibodies and their corresponding Fc and Fab fragments7,8 presented a similar trend in the unfolding events, with the unfolding of the Fab fragment occurring at lower temperatures than that of the Fc fragment.
Using three different humanized IgG1 antibodies we show here that different melting profiles can be observed depending on the stability of the Fab fragment. We also demonstrate that the stability of the Fab fragment is significantly affected by the sequence of the variable domains. Therefore, one cannot assign a priori the first transition observed by DSC of the intact antibody to the unfolding of the Fab fragment; rather it is the experimental enthalpy of unfolding, as determined by the peak area in the DSC thermogram, which may be used as the indication for which transition represents the Fab fragment unfolding. This approach relies on the assumption that the Fab fragment unfolds in a cooperative manner, that is, only one transition is observed in the thermogram of the Fab fragment. If the coupling among the domains in the Fab is disrupted by CDR grafting and humanization process, the thermogram of the Fab fragment may present multiple transitions, and the interpretation of the DSC profile for an intact antibody will become more complex.
As will be shown below, multiple transitions observed in Fab unfolding should be carefully examined because may represent artifacts of papain digestion. One has to emphasize that the amplitude criterion can be used for DSC measurements only, and cannot be extended to other methods which track the temperature-induced unfolding by a spectroscopic signal (like circular dichroism or fluorescence).
One humanized antibody of type IgG1k was produced and purified in Merck Research Laboratories, and will be further referred as monoclonal "Mrk." The other two humanized antibodies, trastuzumab (Herceptin®) and bevacizumab (Avastin®), are produced by Genentech and are commercially available. The latter monoclonals will be further referred to as "Her" and "Ava," respectively. Unless otherwise stated, the proteins were dialyzed prior to DSC measurements against the same buffer containing 10 mM sodium phosphate, 150 mM sodium chloride at pH 6.5. The protein concentration of intact antibodies or Fab and Fc fragments was measured in 6 M Guanidine Hydrochloride (GuHCl) spectrophotometrically. The extinction coefficient at 280 nm was calculated for each protein based on the content of tryptophans, tyrosines, and cystines.13
Monoclonal antibodies were digested with immobilized papain (Pierce kit cat #44885, Rockford, IL) following manufacturer’s instructions, with the exception that the incubation at 37°C was carried overnight. For Mrk and Ava antibodies, the Fab and Fc fragments were separated using Protein A column as described in the kit. Her antibody Fab fragment was found to bind to Protein A (it eluted from protein A column in the Fc fraction) and, therefore size-exclusion chromatography (SEC) was used for fragment separation. SEC was carried out on a ProStar HPLC system (Varian, Walnut Creek, CA) using two G3000SWXL columns (Tosoh Bioscience, Tokyo, Japan) connected together. The mobile phase used was 25 mM sodium phosphate, 150 mM sodium chloride, pH 6.8 and the flow rate was 0.5 mL/ min. With papain digestion carried overnight the digestion of Mrk antibody was not complete. Therefore, the Fc fragment of Mrk antibody was further purified using SEC (the same as above) to eliminate the undigested and partially digested (Fc-Fab) antibody.
Mass spectrometry together with SEC and SDS-PAGE revealed that approximately 75% of the Ava Fab fragment sample contained one or two clips in the VH domain and, importantly, that these clipped molecules are in a dimeric form (data not shown). Ava antibody Fab fragments, intact or clipped, were separated using the SEC system described above. The intact fraction of Ava Fab sample, purified by SEC, had 95% purity. Mass spectrometry confirmed that the intact fraction is formed by an intact light chain and an N-terminal heavy chain fragment that resulted from the expected papain digestion at H/T position in the hinge region, without additional cleavage sites in the VH domain. The Fab fragment from the Ava antibody, containing intact and clipped molecules, was found to have a broad, two-peak transition by DSC, suggesting that there are at least two distinct species with different melting temperatures. Based on the DSC profiles of the purified clipped and intact Fab fragments (data not shown), we propose that the transition with the lower melting point in the dimer(clipped)/ monomer (intact) mixture represents the dissociation/unfolding of clipped monomers. It is possible that the dimers, held together by non-covalent interactions, dissociate at higher temperatures and the clipped monomers are destabilized compared to the intact monomer due to the disruption of the polypeptide chain.
Two milligrams of Mrk antibody was mixed with 2.2 ml of peptide N-glycosidase F (PNGase F, New England Biolabs, Ipswich, MA) and 20 ml of G7 buffer provided with the enzyme. The digestion was carried out for 24 h at 37°C. PNGase F converts the asparagine residue that contains the glycan chain into aspartic acid, and the completeness of the deglycosylation can be monitored by cation-exchange chromatography (CEX). CEX was performed with ProPac WCX-10 4 250 mm column (Dionex Corporation, Sunnyvale, CA). Ten millimolars sodium phosphate pH 6.5 was used as mobile phase A. Mobile phase B had the components of mobile phase A plus 0.5 M NaCl. The antibody was separated with 10-45% gradient B over 25 min at 0.5 mL/min flow rate.
The Fab fragment from Ava antibody sample was separated by SEC on Waters 2690 Separations Module (Waters Corporation, Milford, MA) using a G3000SWXL column (Tosoh Bioscience) with online refractive index (RI) (Waters 2410 RI Detector) and light scattering (LS) detection (PD2000, Precision Detectors, Franklin, MA). The mobile phase was 150 mM sodium chloride at a flow rate of 0.5 mL/min. The ratio of the refractive index and light scattering signal was used to calculate the ratio of molecular weights of the two Ava antibody Fab fragment fractions as described previously.14 Shortly, RI and LS signals were normalized by the height of the putative dimer peak. The LS/RI for the maximum of the monomeric peak and the monomeric peak area were calculated to be 0.53 and 0.49, respectively. Because the LS/RI ratio is proportional to the molecular weight of the species, the putative dimer was found to have twice the molecular weight of the monomer.
SDS-PAGE was performed using Tris-Glycine 4-20% gel (Invitrogen, Carlsbad, CA). Samples were prepared according to manufacturer’s instructions and 15 mg of the Ava antibody Fab fragment were loaded per lane.
Mass spectrometry analysis was performed using a capillary HP1100 HPLC system coupled to an Agilent MSD-ESI-TOF VL mass spectrometer. In order to examine the masses of the light and heavy chains, the Fab samples were incubated with 50 mM DTT for 15 min at 75°C, followed by dilution into formic acid. The samples were injected onto a 0.5 mm50 mm Monolithic PSDVB column (Dionex), equilibrated at 70°C and eluted with a gradient of 0.1% formic acid in water and acetonitrile at a flow rate of 0.02 mL/min. Mass reconstruction from the raw data was accomplished with the Protein Confirmation software provided with the system.
The DSC measurements were performed, unless otherwise mentioned, at 1 mg/mL protein and 1°C/min scan rate on a MicroCal VP-Capillary DSC Platform (MicroCal, LLC, Northampton, MA). The DSC profiles were calculated using the Origin 7.0 software: the buffer background was subtracted and the thermogram was normalized to the molar concentration of the protein. The final excess heat capacity thermogram was obtained by interpolating a cubic baseline in the transition region.
The melting temperatures reported represent peaks in the experimental thermograms and the enthalpy of unfolding was obtained using the Origin 7.0 software by integration of the area under the melting curves in the temperature range 55–90°C.
Because the transitions reported in this study are irreversible, all the experimental values reported for melting temperatures and enthalpies should be regarded as "apparent" values.
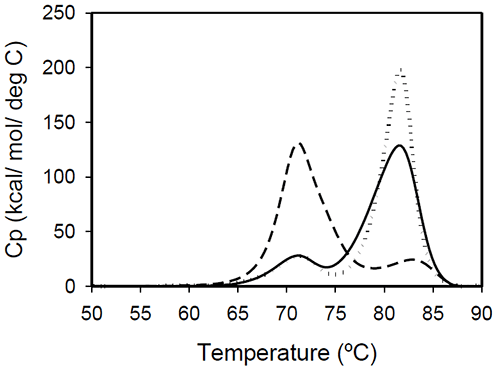
The profiles of temperature-induced unfolding of three humanized IgG1 monoclonal antibodies (Mrk, Her, and Ava), under the same solvent conditions, are shown in Figure 2.
|
The thermogram of each protein consists of two transitions, centered around 70 and 80°C, but with different corresponding apparent enthalpies. For the Ava antibody, the first transition has larger amplitude than the second transition, while for Mrk and Her antibodies, the second transition has the larger amplitude. To evaluate the errors in determining melting temperatures and enthalpies, duplicate measurements for each antibody were performed several weeks apart. The results indicate that the variation of the melting temperature is within 0.25°C and that the variation in the measured enthalpy is less than 3.5%.
The DSC profile observed for the Ava antibody is similar to the temperature induced unfolding of another human IgG111 and a murine IgG2a antibody.7 Two transitions were observed for the murine antibody, with the first peak area about twice the area of the second peak. Further studies with fragments of the murine IgG2a antibody clarified that the first transition corresponds to the Fab fragment unfolding, while the second transition corresponds to the melting of the Fc fragment. Similar trend was reported for a different murine IgG2 antibody and its Fab and Fc fragments:8,15 between pH 6 and 8 the intact antibody presents two transitions, with Fab unfolding corresponding to the first transition and Fc unfolding representing the second transition. In contrast, for a murine IgG1 antibody, only 1 peak was observed at pH 6.0,6 the difference from IgG2a being ascribed to differences in the flexibility in the hinge region between isotype 1 and 2 in mice.
Among the three humanized IgG1s investigated in this study, the temperature-induced unfolding of the Ava antibody seems to follow the pattern observed for the two murine IgG2 antibodies. However, it is obvious that this trend is not followed by other humanized IgG1 monoclonal antibodies and, as it will be demonstrated later, the interpretation of the murine thermogram is not applicable to humanized antibodies. Detailed analysis of Fab and Fc fragments of humanized IgG1 antibodies described below shows a clear distinction between the thermograms of the Ava and murine IgG2 antibodies: the first transition in the Ava antibody represents the unfolding of the Fab fragment and the unfolding of one domain in the Fc fragment, while only the unfolding of the Fab fragment contributes to the first transition of the murine IgG2 antibodies reported so far.
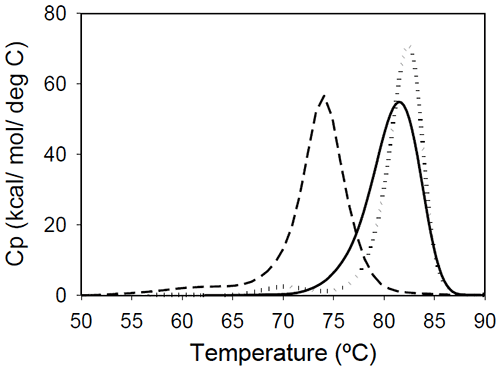
Fab fragments from Mrk, Her, and Ava antibodies were obtained by papain digestion followed by chromatographic separation using protein A and/or size-exclusion columns. The purity of the Fab fragments, assessed by SEC and non-reduced SDS-PAGE was ~90% for the Her antibody, and larger than 95% for Mrk and Ava antibodies (data not shown). Temperature-induced unfolding profiles of Fab fragments from Mrk, Her, and Ava antibodies are shown in Figure 3.
|
The thermograms of Mrk and Her Fab fragments present only one peak around 80°C, the temperature where the large-amplitude transition was observed for the corresponding intact antibodies. For both Mrk and Her Fab fragments the experimental enthalpies of unfolding are in the range 320–340 kcal/ mol. For each fragment, the variation in the measured enthalpy was found less than 3% and the variation in measured Tm<0.2°C.
The Fab fragment from Ava antibody was found to show structural heterogeneity after papain treatment (see Experimental section for more details). Additional papain cleavage sites in IgG1 variable domains were reported and it has been proposed16 that CDR conformation could be the determining factor for the increased susceptibility for enzymatic digestion. The DSC profile of the intact Ava Fab fragment (purified from the heterogenous sample by SEC) is shown in Figure 3 and it has a melting temperature of ~74°C, close to that observed for the first transition in the full-length Ava antibody (Figure 2). The experimental enthalpy of unfolding of the Ava antibody Fab fragment is 316 kcal/mol, within 1% of the experimental enthalpy of the Her antibody Fab fragment. It is worth noting that the clipped Fab fragment of Ava antibody is significantly destabilized compared to the intact fragment and has a strong tendency to dimerize. Based on the concentration dependence (data not shown), the dimers formed upon papain cleavage are distinct from dimers previously reported for the intact Ava antibody.17
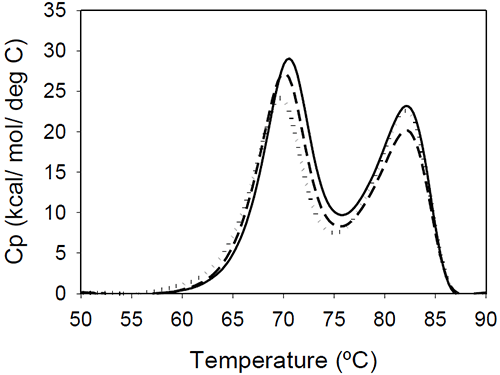
DSC results of the Fc fragments resulting from Mrk, Her, and Ava antibodies are shown in Figure 4.
|
The unfolding presents two peaks centered around 70 and 80°C. The measured enthalpies of unfolding for Mrk Fc are about 190 kcal/mol and 160 kcal/mol, respectively (as found with the intact antibodies and Fab fragments, the variations in the measured enthalpies did not exceed 3.5%). No significant variations among the thermograms are expected, based on the fact that all three humanized IgG1 antibodies have the same amino acid sequence in the Fc fragment resulting from papain digestion. As it will be shown later in this study, the transition of the Fc fragment centered around 70°C is significantly affected by glycosylation, so it may be possible that the small differences in the first peak reflect different glycosylation patterns for the different antibodies.
The bimodal distribution of the temperature-induced unfolding of the human IgG1 Fc fragment is in agreement with previous reports.18,19 Studies using fluorescent-labeled domains12 or Fc fragments with different glycosylation patterns20 indicated that the first transition represents CH2 domain unfolding and the second transition represents CH3 domain melt. The two transitions may be somehow coupled around neutral pHdue to a contact area12 of 400 Å2. The coupling of the unfolding of the CH2 and CH3 domains is suggested also by the DSC profile of a fusion protein containing the Fc fragment.19 In this fusion construct, the melting temperature corresponding to CH2 domain unfolding shifts to a lower value, and the enthalpy of the transition corresponding to CH3 is increased relative to that of CH2.
The ratio of the enthalpies of the two transitions (CH2/CH3) is slightly larger in previous reports (between 1.4 and 1.6), and this may be due to differences in the glycosylation pattern and/or due to differences in solution pH. Studies on rabbit IgG9 showed that, at pH greater than 4, the first Fc transition is larger in amplitude than the second, but at pH 3.5 the second Fc transition becomes about three times larger than the first one. It is interesting to note that the Fc fragments from murine IgG2a or IgG2b isotypes present only one transition, as reported in studies using two different murine molecules.7,15 It may be possible that the coupling between the CH2 and CH3 domains is much stronger in a murine monoclonal antibody compared to the coupling in an human corresponding isotype, and the two domains unfold in a cooperative manner, similar to Fab fragment unfolding.
Temperature-induced unfolding of the Mrk antibody and its Fab and Fc fragments are shown in Figure 5. The variations in measured melting temperatures between intact antibody and fragments are less than 0.5°C, and the relative variation in total measured enthalpy between intact antibody and fragments is less than 10%.
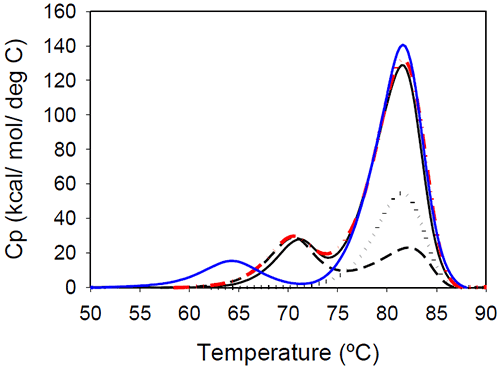
|
Figure 5 also contains a thermogram that was obtained as a sum of 1:2 molar contributions from the Fc and Fab fragments. The calculated thermogram is in good agreement with that obtained the intact antibody. In a separate experiment, a 1:2 molar mixture of Fc and Fab fragments corresponding to 1 mg/mL of the full length antibody was prepared. Results of DSC measurements performed on the Fc/Fab mixture for overlapped very well with measurements performed with the full length, intact antibody (data not shown). All these observations support the conclusion that the unfolding of the Fab and Fc fragments are independent events.
To our knowledge, this is the first report to demonstrate that the thermogram of the intact antibody at neutral pH can be expressed as the sum of the thermograms of the Fab and Fc components. This additivity demonstrates that the temperature induced unfoldings of the Fab and Fc fragments in an intact antibody are independent from each other. Table 1 summarizes the results of DSC measurements on all three monoclonal antibodies and their fragments.
| First Transition | Second Transition | ||||
|---|---|---|---|---|---|
| Protein | Tm (8°C) | Enthalpy (kcal/ mol) | Tm (8°C) | Enthalpy (kcal/ mol) | Total Enthalpy (kcal/ mol) |
| Mrk | |||||
| Intact | 71.0 | 157 | 81.5 | 786 | 943 |
| Fab | 81.4 | 341 | 341 | ||
| Fc | 70.6 | 192 | 82.0 | 158 | 350 |
| Her | |||||
| Intact | 71.4 | 163 | 81.8 | 835 | 998 |
| Fab | 82.4 | 318 | 318 | ||
| Fc | 69.7 | 170 | 82.2 | 155 | 325 |
| Ava | |||||
| Intact | 71.2 | 800 | 82.9 | 138 | 938 |
| Fab | 74.1 | 316 | 316 | ||
| Fc | 70.0 | 185 | 82.0 | 138 | 323 |
| The separation between the first and second transition was determined by the peack valley in Cp. | |||||
For Her and Ava monoclonal antibodies, the relative variations in total measured enthalpy between intact antibody and fragments are 4% and 2%, respectively. However, the variations in the apparent melting temperatures between the intact antibody and fragments exceed, in some cases, 1°C. Consequently, for Ava and Her monoclonal antibodies, the agreement between the calculated thermogram from those of the fragments (2*Fab+Fc) and the experimental thermogram of the whole antibody is not as good as in the case of Mrk antibody. It may be possible that the impact of the irreversibility of unfolding is different from protein to protein. As it will be discussed in more detail later, it was found that the experimental melting temperatures, and not the measured enthalpies, show larger variations when the scan rate is changed in a thermal melt. The independence of Fc and Fab fragments unfolding assessed by calorimetric measurements was reported for a rabbit antibody at pH 3.5,9 however, extrapolation of this result to neutral pH is not trivial for the following reasons: (1) the antibody adopts an alternative conformation at low pH, with significant changes in the coupling among different domains; (2) the reversibility of temperature-induced unfolding varies dramatically with pH, from a completely reversible process at pH 2 to a completely irreversible process at neutral pH.5 An important consequence of the demonstrated uncoupled thermal unfolding of the Fab and Fc fragments is that any findings pertinent to the stability of the Fc or Fab fragments should be relevant for the stability properties of the intact antibody as well.
Glycan removal from the CH2 domain of the full-length Mrk antibody resulted in a shift of the first transition (Figure 5), leaving the transition that occurs at the higher temperature unchanged. This suggests that the first peak in the thermogram of the full-length Mrk antibody represents the unfolding of the CH2 domain. Similar findings were reported for other human Fc fragments:20 only the first peak shifted upon changes in glycosylation, while the second peak was glycosylationindependent.
Changes in the glycosylation pattern may not affect the intrinsic stability of the CH2 domain, but instead diminish the stabilization energy for that domain resulting from CH2-CH2 and/or CH2-CH3 inter-domain interactions. Previous studies on unglycosylated, murine CH2 domain demonstrated that this domain is relatively unstable compared to other monoclonal antibody domains and the lower instability was proposed to be due to a higher flexibility of the structure needed to accommodate interactions with different Fc receptors.21 The unfolding of intact, full-length monoclonal antibodies at pH 6.5 is completely irreversible. Samples that were heated up to 90°C do not show any unfolding transitions when reheated in a second scan. This is in agreement with previous reports showing that the unfolding of monoclonal antibodies is irreversible around neutral pH and reversible at pH 2 where the protein is in an "alternative immunoglobulin" conformation.5 For a rabbit antibody at pH 3.5 the additivity of Fc and Fab unfolding was also demonstrated.9 Our data presented above shows similar additivity for a humanized monoclonal antibody at pH 6.5, in a conformation distinct from the acid-induced folded form. For a murine monoclonal antibody a difference of ~80 kcal/mol (about 10%) between the enthalpy of the intact antibody and the sum of enthalpies of fragments unfolding at neutral pH7 was reported.
Although differences in the measured enthalpies of unfolding and/or melting temperatures between intact monoclonal antibodies and fragments may be considered an indication that the fragments do not unfold in an independent manner (coupling between Fab and Fc unfolding was proposed for a murine monoclonal antibody)15 caution should be exerted when interpreting irreversible melts. It may be possible that a correct evaluation of the area for the transitions that occur at high temperatures may be precluded by different contributions of the aggregation process to the thermogram. The rate of aggregation depends on a multitude of factors, including sequence propensity for aggregation, protein concentration, solvent conditions, and, importantly, configuration of the calorimeter cell. Consequently, the contribution of aggregation/precipitation at high temperatures may vary from case to case and, for some monoclonal antibodies, obtaining post-transition baselines at high temperatures may be impossible in a non-capillary DSC.
The influence of aggregation on the DSC profile for a human IgG1 was demonstrated by studies using an antibody against human tumor necrosis factor-α (TNF-α). In that particular case, the Fab fragment had a melting temperature close to that of the CH2 domain (similar to the Ava case), but the transition for the CH3 domain unfolding in the intact antibody was obscured by extensive aggregation.22 As a result, the apparent DSC profile for the human IgG1 antibody against TNF-α has only 1 peak. According to the calorimetric enthalpies reported,22 adding the contributions of Fab and Fc fragment enthalpies (310 kcal/mol for Fab, 198 kcal/mol for CH2 and 165 kcal/mol for CH3), one would expect about 983 kcal/mol for the full-length antibody. However, only 678 kcal/mol were reported for an "intact" molecule, so the enthalpy balance clearly indicates that a significant part of the Fab and Fc unfolding is not captured in the profile for the "intact" antibody.
Based on our experience, the 2-peak distribution of the DSC profile of IgG1 monoclonal antibodies is a shape to be expected in most cases studied, and it reflects the fact that the melting point of the Fab fragment is either comparable to that of the CH2 domain (when the first transition of the intact, full-length antibody has the larger area) or to that of the CH3 domain (when the second transition of the intact, full-length antibody has the larger area). However, it may be possible that the Fab fragment melting temperature occurs at distinct values which are lower, intermediate, or higher than those of the domains in the Fc fragment. For those cases, it is expected that the DSC profile for an IgG1 antibody will present 3 peaks.
The relative shift of the melting temperature for the Fab fragment compared to the Fc fragment may be produced by either affecting Fab stability (by CDR grafting and humanization process) or Fc stability (by changing the pH). For the Ava antibody, changing the pH from 6.5 to 5.5 produces a separation of the CH2 and Fab unfolding, and the DSC profile changes from a two-peak to a three-peak shape, with Fab melting occurring at intermediate temperatures between those of the CH2 and CH3 domains (Figure 6A). The temperature-induced unfolding of Mrk antibody at pH 5, 6, and 7 is shown in Figure 6B.
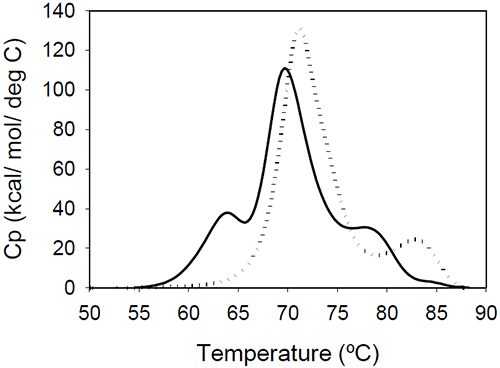
|
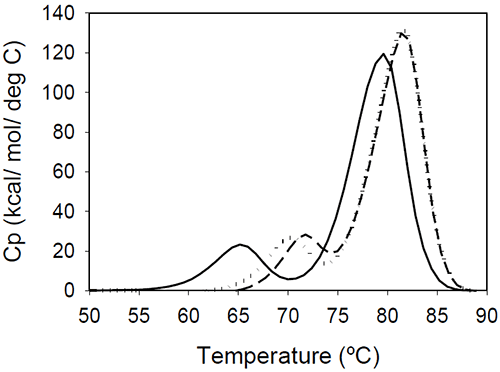
|
It is evident that the pH has a greater impact on the melting temperature of the first transition than on the melting temperature of the second transition. Although the 3-peak model seem to suffice to describe the examples above and the data published in the literature, we cannot exclude a priori that more complex patterns may be observed for some humanized IgG1 monoclonal antibodies. In those cases, more investigations will be required to explain the profile and we speculate that the deviations may be a consequence of significant structural heterogeneity or may reflect the impact of CDRs on the disruption of the cooperative (simultaneous) unfolding of the variable domains.
Our results demonstrate that solvent-exposed loops of CDRs and humanization- related mutations have a significant impact on the stability of the Fab fragment and the impact can be much larger than the differences in stability between different human Fab frameworks. The stabilities of Mrk and Her Fab fragments are comparable, but both are significantly different from the stability of the Ava Fab fragment, despite the fact that there is a greater sequence similarity between Ava and Her than between Mrk and Her antibodies. Ava and Her antibodies contain the same consensus frameworks, VHIII for the heavy chain and Vk1 for the light chain.23,24 The Mrk antibody was humanized using different human frameworks, VHII for the heavy chain and Vκ2 for the light chain. The light and heavy chain frameworks used in Ava and Her molecules are among the most stable domains of different families (2.1 and 3.0 kcal/mol, respectively), while the light and heavy frameworks used in the Mrk antibody are among the least stable domains (1.5 and 1.6 kcal/mol, respectively).4 Sequence alignment of the variable domains in Her and Ava antibodies is shown in Figure 7.

|
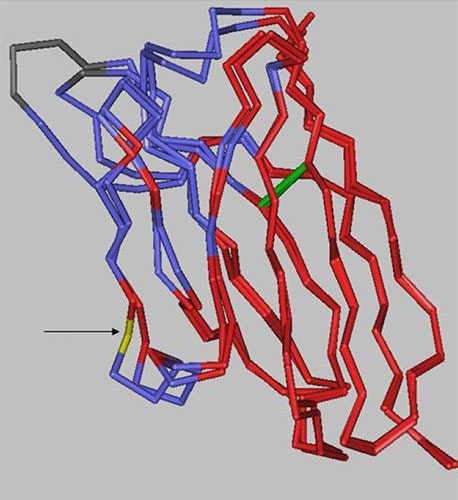
There are 10 positions in the heavy chain and 2 positions in the light chain outside of the CDRs that are different between the two antibodies. However, as the structural alignment of the heavy chain variable domains shows (Figure 8), the changes are limited to solvent-exposed regions, leaving the immunoglobulin fold core unaltered.
|
Previous studies on multi-domain proteins25,26 modeled the stabilization due to the interactions between domains by inclusion of an interface free energy. This free energy goes to zero after the unfolding of one of the domains involved in the pair wise interaction, so the stabilization is observed only for the least stable domain, that is, the domain with the lowest melting temperature. Therefore, the stability of an antibody can be affected by CDR grafting and the humanization process by a combination of any of the following factors: (1) selection of human variable framework; (2) impact of CDRs and additional mutations in the human framework on the intrinsic stability of each variable domain (VH or VL); (3) impact of CDRs and additional mutations in the human framework on the energy of interaction between the domains. It is beyond the goal of this study to determine whether the significant differences in the stability between Ava and Her Fabs are mainly due to changes of intrinsic stabilities of the domains or to significant modifications of domain/domain interactions. Although mutations in key positions of human germline consensus sequences can significantly alter the stability of variable domains,27 we speculate that the differences between Ava and Her stability are likely due to changes in domain–domain interactions. Previous studies with scFv fragments revealed that CDR3 from both VH and VL have an important role in the interaction energy between the variable domains.4 The stabilization energy due to interactions between domains can be regarded as a factor that increases the melting temperature of the least stable domain. There are several scenarios that can describe the folding of two interacting domains, depending on the intrinsic stability of each partner and the absolute value of the stabilization energy due to the interactions25; one possibility is that two interacting domains unfold in a cooperative manner at a temperature that is higher than any of the melting temperatures observed for the domains when they unfold separately. For the Fab fragment, the interaction energies are significant not only between VH and VL, but also between CH1/CL and between (VH/VL)/(CH1/CL).28 Therefore, the overall stability of the Fab fragment is significantly enhanced compared to the sum of the intrinsic stabilities of participating domains.
Domain–domain interactions may play a dominant role in the stability of the variable domains; however, the impact of CDR grafting and humanization-related mutations on the intrinsic stability of the domains should not be overlooked. Strickler et al.29 challenged the notion that charged residues that are very well exposed to the solvent at the protein surface do not contribute to the stability of the protein. The group developed a computer model using short- and long-range interactions to evaluate the contribution of each residue to the overall protein stability. Redesigned proteins with modified surface charge distribution presented a significant increase in stability, supporting the strategy of protein design by optimization of surface charge–charge interactions.
Because the temperature-induced unfolding transitions presented above are completely irreversible, one may challenge the relevance of the quantitative aspect of the results presented in this study. Our findings, empirical in nature, suggest that as long as the aggregation process following unfolding does not impact significantly the post-transition baseline, the determination of the enthalpy of unfolding can be done with a relatively high precision. Additional observations supporting this statement for the particular case of the unfolding of monoclonal antibodies monitored in a capillary DSC are: (1) the experimental enthalpy of unfolding was found independent of the scan rate (range varied 30–120°C/h); (2) for Mrk antibody, the DSC profiles were independent of protein concentration in the range 0.25–1 mg/mL; (3) the measured enthalpy of the intact antibody unfolding matched the sum of the enthalpies of independent fragments. Because the experimental enthalpy of unfolding is proposed here as the key parameter to identify the unfolding event corresponding to the Fab fragment, the quantitative aspect of this approach may hold as long as the contribution of aggregation process to the thermogram is small. There are other examples in the literature demonstrating good accuracy in the determination of the enthalpy for unfolding events with different degrees of reversibility30 when the aggregation does not significantly alter the post-transition baseline. The irreversibility of the process may have a larger impact on the determination of the melting temperatures. The apparent Tm of the second transition for the Mrk antibody was found to shift with about 3 degrees when the scan rate was varied between 30 and 120°C/h. It is not clear whether this kinetic factor is related to the rate of aggregation and/or may contain also the contribution from the kinetics of intrinsic protein folding or unfolding rates. There are few examples of quantitative analysis of thermograms with a model composed of a two-state reversible unfolding reaction followed by an irreversible process of aggregation.31–33 We did not attempt to use that model because it is obvious that a two-state reversible equation followed by an irreversible step cannot describe the process of multi-domain unfolding of a monoclonal antibody.
Although convincing arguments were presented in several instances when irreversible thermograms were analyzed using thermodynamic models, the applicability of this approach to other irreversible systems should be evaluated with caution.34 In a previous report of DSC study on a murine monoclonal antibody7, a multi-state thermodynamic model was used to analyze the thermograms over a pH range where the reversibility of the unfolding varied from 100% to 0%. The consistent titration curves reported suggest that the analysis may provide a reasonable approximation, although, in principle, the applicability of thermodynamic models may be questioned. In our study, the slight dependence of the apparent Tm on the scan rate indicates that the system is not quite "at equilibrium" even though the contribution of the aggregation may be considered negligible throughout most of the transition. Therefore, no multi-state analysis using thermodynamic models is presented on the monoclonal antibodies and their fragments.
Understanding the stability of monoclonal antibodies is important in the development of new therapeutic agents, and we hope that the results of this study will provide a useful tool for the interpretation of temperature-induced unfolding of IgG1 monoclonal antibodies.
We would like to thank Dr. Henryk Mach for insightful discussions, and Dr. Li Shi for his critical review of the manuscript. Bei Wang was kind to provide technical assistance in operating the calorimeter, and we’d like to acknowledge the contributions of Mark Schaefer and Shiyi Wang to the development of the papain digestion and PNGase F deglycosylation assays.