The combination of dynamic light scattering (DLS) with Raman spectroscopy provides the ability to extract a wealth of chemical, structural, and physical information about biotherapeutic proteins under formulation conditions.
Dynamic light scattering (DLS) using noninvasive backscatter (NIBS) detection technology is capable of measuring the hydrodynamic radius of proteins over a large concentration range (0.1 mg/mL to 100 mg/mL). As a result, it is possible to determine, in actual biopharmaceutical formulations, the bulk viscosity/restricted diffusion interaction parameter (kD), particle interaction parameter (B22), melting temperature, onset temperature of protein aggregation and transition enthalpies to assess colloidal stability.
The unique coupling of DLS with Raman spectroscopy provides the ability to simultaneously correlate colloidal parameters to protein structure under a variety of stress conditions, e.g., thermal, formulation, chemical degradation, extrinsic particulates, to enhance our understanding of protein therapeutic formulations.
Dynamic light scattering (DLS) is a workhorse technique for determining the hydrodynamic diameter of biotherapeutic proteins in solution, sample polydispersity, and sample interactions. Raman spectroscopy derives information about protein secondary and tertiary structure by monitoring molecular vibrations. By combining these two analytical approaches into a single system, a wealth of chemical, structural and physical parameters can be determined. Protein secondary and tertiary structure, melting temperature, onset temperature of aggregation and transition enthalpy values can all be derived, as well as aggregation propensity, protein solubility, and the potential for high viscosities at formulated concentrations. The complementary nature of the results obtained from DLS and Raman spectroscopy on the same sample may provide unique insights into the mechanisms of aggregation and unfolding. Here we describe the utility of DLS to aid in the understanding the colloidal stability of protein therapeutics.
Malvern Instruments’ Zetasizer Helix (ZS Helix) integrates a fiber-coupled Raman spectrometer with a Zetasizer Nano ZSP to provide DLS (colloidal stability) and Raman (conformational stability) data sequentially on a single sample. The Zetasizer Nano system integrates proprietary non-invasive backscatter (NIBS) detector technology with dynamic (DLS), static (SLS) and electrophoretic (ELS) light scattering to measure the hydrodynamic radius of proteins from 0.15 nm - 5 µm, from 0.1 mg/mL to ≥ 100 mg/mL. Raman spectra are collected using 785 nm excitation (~280 mW) from 150 - 1925 cm-1 at 4 cm-1 resolution. Sample aliquots (~120 µL) are placed into a 3 mm quartz cuvette and positioned in a temperature-controlled compartment that provides temperature control from 0°C - 90°C ± 0.1°C. Thermal ramp studies are conducted by collecting Raman and DLS data over a series of pre-defined 0.1°C - 5°C step-wise increments. Isothermal incubation studies are conducted by collecting a series of Raman and DLS data over a predefined time interval at a desired temperature set-point.
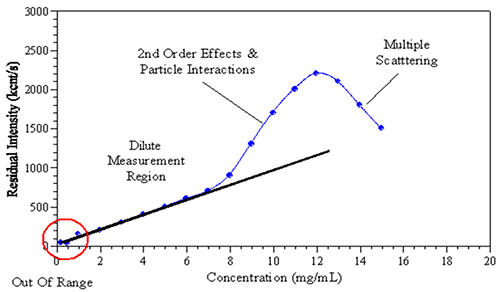
The formulation of monoclonal antibodies at high concentration has drawn great attention within the biopharmaceutical industry in recent years. However, for DLS, size measurement at formulated concentrations was limited by multiple experimental factors. As shown in Figure 1, increasing sample concentration could affect the observed scattering intensity and consequently the size determination. Factors affecting the measurement could include: multiple scattering, viscosity, positive/negative virial effects, and self-association. Deconvolution of the above effects allows for interpretation of high-concentration sample results.
|
To better understand the effects of the aforementioned factors on the DLS data, it is convenient to consider an individual effect in isolation. Table I (below) summarizes the expected changes with increasing sample concentration to the reported size, polydispersity, and correlation function, as well as established correction methods, for each of the unwanted factors.
| Factors | Apparent Size | Apparent Polydispersity | Correlation Intersection | Correction Method |
|---|---|---|---|---|
| Multiple Scattering | Decrease | Increase | Decrease | Backscattering |
| Restricted Diffusion | Increase | No Change | No Change | Viscosity Correction |
| Negative B22 | Increase | Increase | No Change | No |
| Positive B22 | Decrease | No Change | No Change | No |
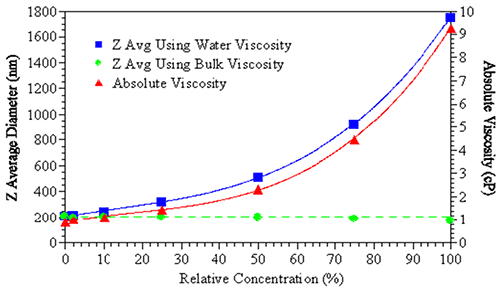
Figure 2 (below) demonstrates the effect of viscosity correction on the reported size of a sample. Using the viscosity of water as an estimate of the sample viscosity results in an increase in apparent size of the suspended particles (blue squares). However, when the true bulk solution viscosity is used to correctly fit the DLS results, no apparent size change is observed (green circles). Particle interaction terms including attractive/repulsive, B22 (second virial coefficient) or kd (diffusion interaction parameter) could be indicated and will be covered in the following section. It should be noted that for virial effects, there is currently no agreed approach to correct DLS data.
|
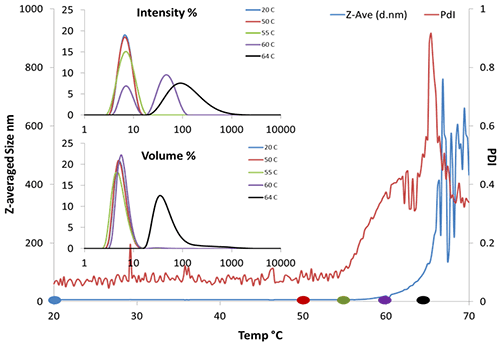
Static light scattering (SLS) or turbidity coupled with thermal ramping have been routinely used as rapid formulation screening tools for protein stability. Essentially, these methods measure the aggregation onset temperature Tonset. DLS could also be utilized in this fashion. Figure 3 shows the aggregation onset for BSA is approximately 62°C by Z-averaged size. Additionally, as long as the correlogram remains valid, DLS offers the size distribution (Fig. 3, insert) and polydispersity (PDI), where SLS offers only the intensity. The size distribution trend shown here clearly reveals the appearance of a second peak at 90 nm near 60 °C. The onset temperature from PID could be as early as 55 °C, indicating aggregation may begin even earlier.
|
|
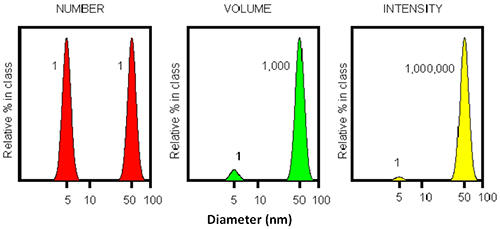
The ability to detect trace amounts of aggregates stems from the inherent sensitivity of DLS. Figure 4 demonstrates that for a simple mixture of two particles with an order of magnitude difference in diameter, the larger 50 nm particle will show a scattering intensity a million times stronger than the smaller 5 nm particle. This infers that even a single large particle mixed with a million small particles results in an equal detected intensity! Thus, DLS could be useful as the earliest detector for the appearance of aggregates. However, it should be noted that the resolution of DLS is inherently limited to a factor of three in size.
Though Tonset might be sufficient for initial formulation screening, no understanding of protein interaction can be reached from such experiments, let alone the prediction of protein stability. Recently, more attention has been focused on investigating intermolecular parameters as predictors of protein aggregation kinetics, which have been successfully correlated with B22 and kD [1]. A large positive value of B22 indicates that solutes prefer association with the solvent over self-association, either due to strong electrostatic repulsion between solute molecules or positive interaction between solute and solvent. In contrast, large negative values of B22 indicate strongly self-attractive systems, where the particles prefer self-association or aggregation over complete solvation.
In the case of DLS, kD can be measured directly and is related to B22 by the following equations [2]:
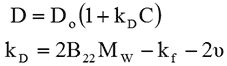
Here, C is the sample concentration and D refers to the diffusion coefficient measured by DLS. D0 is the self-diffusion coefficient (the value of D at zero concentration). Mw is the sample molecular weight, kf is the first order concentration coefficient of the friction and ν is the partial specific volume of the solute.
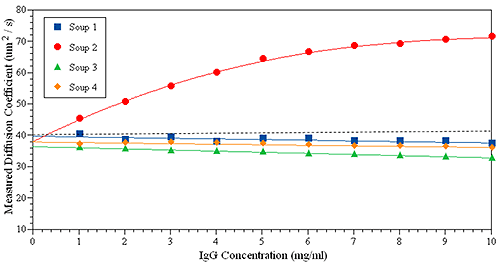
The following experiments demonstrate how to apply kD to predict the protein stability for an IgG in four formulations. In Figure 5 only Soup 2 (red circles) has a positive kD, indicating a strong repulsive interaction between the protein samples. The other three samples show slightly negative kD. It should be mentioned that large positive kD usually indicates strong repulsive interactions, or positive B22. However, when kD is close to zero it becomes difficult to directly link to B22.
|
|
SLS can be measured at a fixed angle for B22, and zeta potential measured to provide relative charge, as summarized in Table 2. Putting all of the information together, a comprehensive profile of protein properties can be obtained to predict protein stability. For example, we would predict that formulation 2 is the most stable formulation for this IgG, while formulation 1 is the least stable.
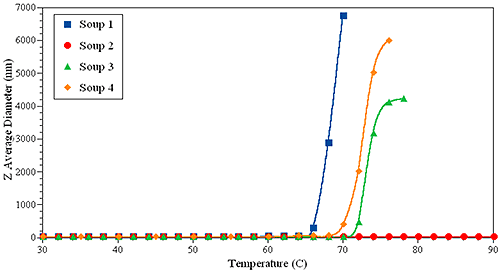
To support this prediction, thermal ramping experiments were performed on the same set of samples, and are shown in Figure 6. Interestingly, sample 1 has the lowest Tonset, close to 66 °C, while formulation 2 is the most stable, showing no evidence of large aggregates up to 90 °C.
| IgG Formulation | B22 (10-5ml mol/g2) | kD (ml/g) | Charge |
|---|---|---|---|
| Soup 1 | -1.5 | -5.2 | 3.3 |
| Soup 2 | 127.5 | 31.9 | 9.7 |
| Soup 3 | 10.4 | -9.7 | 5.1 |
| Soup 4 | 2.3 | -4.7 | -2.8 |
The combination and integration of DLS and Raman spectroscopy into a single instrumental platform provides unique analytical capabilities to determine protein secondary and tertiary structure, melting temperature, and size and onset of aggregation for the same sample, without altering the testing conditions. Here we have described the unique ability of DLS, when combined with noninvasive backscatter detector (NIBS) technology, to derive the bulk viscosity/restricted diffusion interaction parameter (kD), particle interaction parameter (B22), melting temperature, onset temperature of aggregation and transition enthalpies on high concentration formulations. A subsequent study describes the unique spectra-structure properties that may be correlated using Raman Spectroscopy (Understanding the Conformational Stability of Protein Therapeutics with Raman Spectroscopy). Future studies will report on the unique correlations that may be derived from the combination of DLS and Raman spectroscopy to improve the understanding of the kinetics, thermodynamics and mechanism(s) of aggregation and the correlation of specific protein structural motifs to aggregation to improve product knowledge and formulation stability.
Malvern Instruments' Bioscience Development Initiative (BDI) was established to accelerate innovation, development and the promotion of new technologies, products and capabilities to address unmet measurement needs in the biosciences markets.