See how DSC can be used to assess the effects of polyol excipients on antibodies. Differential scanning calorimetry was used to monitor polyol-induced increases in thermal stability across seven monoclonal antibodies. The increases in thermal stability were measured as a function of polyol concentration and orientation of hydroxyl groups. It was found that increases in thermal stability were linear with respect to alcohol concentration. Further, the increase in thermal stability was dependant on the transition temperature; antibodies with lower transition temperatures exhibited a greater increase due to the addition of polyols than antibodies with a higher transition temperature.
Aggregation of proteins has been an intense area of study largely due to the growth of the biotechnology industry. As protein therapeutics are developed, aggregation must be monitored and reduced as it can potentially cause immunogenicity1 as well as a possible decrease in activity.2,3 One of the ways aggregation is controlled is through the use of osmolytes as excipients.4 Many excipients have been shown to induce stability through preferential exclusion. A particular subclass of these molecules includes polyols and carbohydrates.5,6 Polyols are typically added to protein formulations in order to increase tonicity and improve stability.
The addition of polyol osmolytes has been shown to prevent aggregation, retain activity, inhibit chemical degradation, enhance the refolding of proteins,7 and increase the thermal stability of proteins.4-6,8-11 It has been shown that the more hydroxyl groups on the carbon chain of the polyol, the greater the thermodynamic stability that can be induced. This has been attributed to an entropic effect where the polyol is excluded from the surface of the protein. The polyol has a greater impact on the water-peptide interactions than it does on the interactions with the nonpolar groups that are exposed when the protein is denatured.12
To date, no systematic study has been performed using multiple proteins to address whether different stereoisomers of a polyol would induce more or less thermal stability on a protein. The goal of the study was to understand how the size of a polyol and orientation of the hydroxyl group influence the amount of thermal stability that could be induced upon seven monoclonal antibodies of different isotypes. In order to monitor thermal stability, differential scanning calorimetry (DSC) has been employed. DSC has been used to monitor the unfolding of proteins due to heat denaturation in order to study the unfolding of the domains of an antibody,13 monitor thermal reversibility,14-16 and screen formulations for excipients. 17,18
Seven monoclonal antibodies of varying isotypes were obtained. Two antibodies were κIgG1 (K1A and K1B); two were λIgG1 (L1A and L1B); two were κIgG2 (K2A and K2B); and one antibody was an κIgG4 (K4). Histidine, glycerol, erythritol, ribitol, xylitol, mannitol and sorbitol were obtained. For thermal denaturations, a high throughput DSC equipped with an auto-sampler was used.
Each antibody was dialyzed against RODI water. Stock solutions of histidine, glycerol, erythritol, ribitol, xylitol, mannitol and sorbitol were prepared. A liquid handling robot was used to prepare antibody solutions of 1.25 mg/mL in 10 mM histidine at pH 6.0. A titration was performed of 0-350 mM polyol concentration. Buffer blanks were also prepared.
Each sample was subjected to heat denaturation by scanning the samples and their buffer controls from a temperature range of 30 to 95°C at 60°C/hr. The thermograms were baseline-subtracted and deconvoluted using the software provided by manufacturer to obtain the thermal midpoints (Tm) of each transition. Each antibody was fit to three or four transitions depending on how well the unfolding of the Fab and Fc domains were resolved from each other. Figure 1 depicts the three unfolding transitions of L1B where the CH2 domain and one of the Fab domains are not resolved.
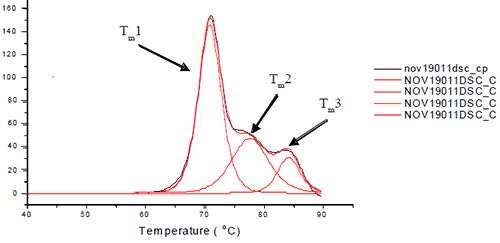
|
The thermograms for each antibody formulated with and without polyol were examined. It was found that the addition of the polyol caused an increase in the Tm value of the molecule. The thermal midpoints (Tm) of each protein were plotted against the polyol concentrations. It was found that there was a linear increase in thermal stability with respect to polyol concentration. An example is shown in Figure 2.
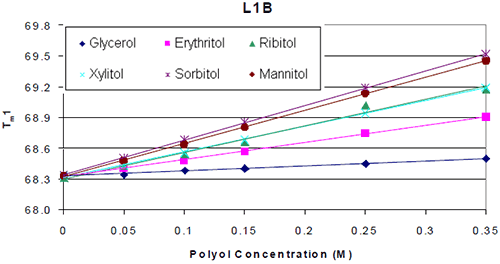
|
The increases in thermal stability were most apparent in the mannitol and sorbitol containing formulations, followed by ribitol and xylitol, then erythritol and finally glycerol. There was no significant difference in the thermal stability induced by the different stereoisomers. Similar trends were seen across all seven antibodies tested. The increase in Tm per mole of polyol (slope of Figure 2) was examined with respect to protein domain (Figure 3a). For K4, the two Fab domains as well as the CH3 domain appeared to exhibit a similar increase in Tm with respect to polyol concentration. There were no significant differences observed in the thermal stability induced by the different stereoisomers of a polyol. This was consistent with the other six antibodies studied.
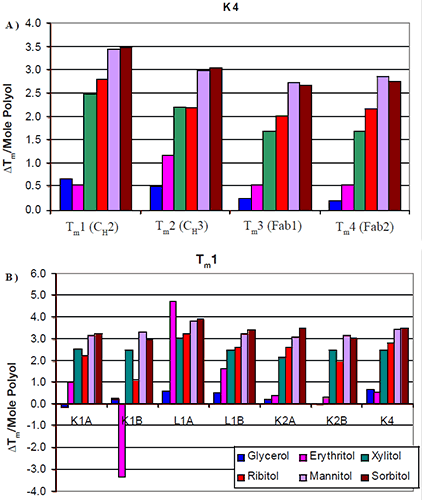
|
The data suggest that a specific stereoisomer of a polyol will not induce more thermal stability than another on a specific domain of an antibody. Since there was no significant difference between the domains, in response to the increased level of polyol and subsequent increases in Tm, the increases in thermal stability were also examined with respect to isotype (Figure 3b). The first Tm for each of the antibodies was compared. All seven antibodies appeared to exhibit a similar increase in Tm with respect to mannitol and sorbitol concentration. Slight differences were observed between xylitol and ribitol for the κIgG1 antibodies. For these two antibodies it appeared as though the increase in thermal stability was not as great in the presence of ribitol as it was in the presence of xylitol. The increase in thermal stability with respect to erythritol concentration varied. In general, the addition of erythritol resulted in less of an increase in thermal stability than ribitol and xylitol but a greater degree of thermal stability than glycerol. For example, erythritol appeared to also have some unique interactions. In contrast with the K1B antibody, it caused a decrease in thermal stability. With the L1A antibody, it caused an increase in thermal stability that was greater than the increase imparted by either mannitol or sorbitol. This suggests that the mechanism of erythritol stability may be different from that of the larger polyols.
The data from all seven antibodies were combined into a single plot (Figure 4a) where the change in Tm per mole of polyol (slope of Figure 2) was plotted against the initial Tm of the 0 mM polyol control (intercept of Figure 2).
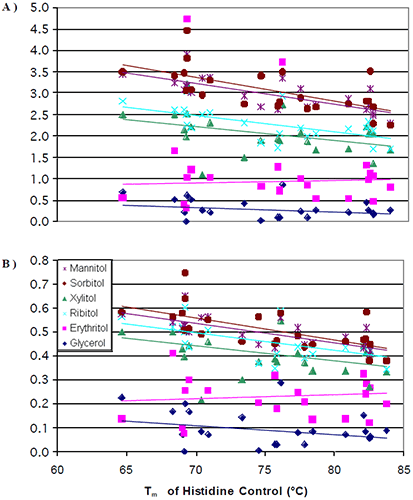
|
It was observed that at lower initial transition temperatures, the amount of thermal stability that could be induced from the addition of a polyol was greater than the amount of thermal stability that could be induced at higher initial transition temperatures. Once again, there were no significant differences in the thermal stability induced by the different stereoisomers. The data was next normalized for the number of hydroxyl groups on the polyols. When this was done, the differences observed for the increase in thermal stability induced by mannitol/sorbitol as compared to xylitol/ribitol started to diminish (Figure 4b). This was to be expected based of the results from the literature.8,12
Two questions remain to be answered: 1) Why is the degree of thermal stability that could be induced by glycerol and erythritol still far less than that of the other polyols examined? 2) Why is it that the proteins with an initial, lower Tm exhibit a greater increase in thermal stability by the addition of a polyol? Polyol osmolytes have been shown to decrease the stability of the denatured state through preferential exclusion.5,6,11,12
Liu and Bolen showed that amino acid side chains had a slight preference to interact with osmolytes as compared to water. The destabilization of the denatured state was attributed to the peptide backbone greatly preferring to interact with water as compared to the osmolytes.19 If interaction between the protein backbone and polyol osmolytes is the deciding factor in determining to what degree the denatured state is destabilized, how can the discrepancies between erythritol and the other polyols be explained? Since all of the polyols have similar structures, it could be argued that they have a similar ability to order water. The real difference between the compounds is their size. It is our hypothesis that the smaller polyols, with less steric hindrance, are able to interact to a greater degree with the peptide backbone, resulting in a more modest increase in thermal stability with the smaller polyols. The smaller polyols are excluded to a lesser degree from the peptide backbone.
The hydrophobic effect and hydrogen bonding are considered two of the dominating forces in protein folding. The proteins that exhibited lower initial Tms (before the addition of any polyols) probably have a less compact structure with weaker hydrophobic and hydrogen bonding interactions. Upon the addition of a polyol, the conformation of the protein will compact such that the peptide backbone will become less exposed to the polyol. The protein structure can only become so compressed. The proteins with the lower initial Tms have the ability to compress further and receive a greater increase in thermal stability through the addition of a polyol.
The results confirm the findings of Back, Oakenfull, and Smith8 as well as Gekko and Morikawa12 where it was observed that there was a linear increase in Tm with respect to alcohol concentration. The larger polyols can influence the degree of stability conferred on the protein as they have more alcohols per molecule. By performing this study on a set of seven antibodies under identical testing conditions, it confirms that the mechanism of stabilization is not protein specific. The findings of greatest interest concern proteins with a lower initial Tm exhibiting a greater polyol-induced increase in Tm than proteins with a higher initial Tm. The antibodies with the higher initial Tms probably have a more compressed, rigid structure with stronger hydrophobic and hydrogen bonds. These proteins cannot benefit as greatly from the addition of a polyol. Proteins with lower initial Tms probably have weaker intramolecular bonds and a more flexible structure. Through the addition of the polyol, the peptide backbone has more room to compress to avoid contact with the polyol,19 and the hydrophobic and hydrogen bonds are now strengthened to a greater degree than they would be in proteins with higher initial Tms.
The authors would like to thank Angela Kantor for all of her support and edits. We would also like to thank Steve Raso, Cliff Entrican and Kyle Wong for their work on assigning domains to the transition temperatures.