This application note describes a new method for the characterization and solubilisation of hydrophobically ion paired enzymes in fluorinated solvents. We report Dynamic Light Scattering (DLS) data obtained using the Malvern Zetasizer ® Nano S™. This technique was used as a tool to determine the particle size of an enzyme-surfactant complex. We used the enzyme Candida rugosa lipase, perfluorinated surfactants Krytox FSL™ and KDP 4606 from Dupont, and perfluoromethyl-cyclohexane as the solvent.
The fluorous biphasic system (FBS) is widely recognized as an environmentally friendly process due to the possibility of simple separation of products from reactants and catalysts hence avoiding complicated and time consuming procedures, such as chromatography. The fluorous biphasic reaction was first reported by Horvath and Rabai [1]. The concept is based on the limited miscibility of partially or fully fluorinated solvents with non-fluorinated compounds. At room temperature, a biphasic system is observed and on warming to above 30°C a homogenous mixture is formed in which reactions can occur. On cooling, the phases partition and products solubilized in the hydrocarbon solvent can be easily separated from spent reagents and catalyst remaining in the perfluorocarbon solvent. It is known that enzymes are capable of catalyzing reactions in a FBS as first reported by Beier and O'Hagan [2] even though the enzyme is insoluble in this system. This research is focused on the potential for biocatalysis in fluorinated solvents using a solubilized form of the enzyme. We apply Hydrophobic Ion Pairing (HIP) methodology to fully solubilize enzymes in perfluoromethylcyclohexane (PFMC) by means of ion pairing to a perfluorinated surfactant. The nature of the enzyme complex can be characterized by various techniques. We report the use of dynamic light scattering (DLS) as a tool to determine the diameter of the particles produced in PFMC.
DLS is a technique that measures the time-dependent fluctuations in the intensity of scattered light from a solution of particles undergoing random, Brownian motion. Analysis of these intensity fluctuations allows for the determination of the diffusion coefficients, which in turn yield the particle size and determine if true HIP as described in Figure 1 is occurring.
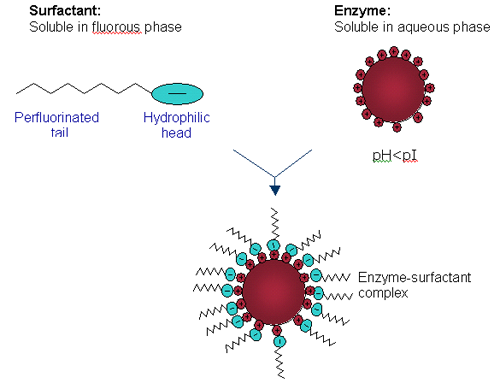
|
Candida rugosa lipase (CRL) is solubilized in PFMC by means of a hydrophobic ion pairing (HIP) mechanism as shown in Figure 1. CRL is dissolved in an aqueous buffer at a pH below the isoelectric point (pI) of the lipase so that all the basic amino acids are protonated. An anionic perfluorinated surfactant, either Krytox (mw ~2500) or KDP (mw ~1400), is dissolved in PFMC. A biphasic system is observed and on stirring, the basic amino acids on the surface of CRL, ion pair to the anionic head groups of the surfactant forming a complex, hence extracting CRL into the fluorous phase. Enzyme concentration in the fluorous phase is determined by UV/Vis spectroscopy at 280 nm.
CRL samples were prepared as follows. The lipase was dissolved in 20mM sodium acetate buffer, pH 3.8 in its native state and its corresponding Krytox and KDP complexes were made by extraction into PFMC as described above. Solutions of each surfactant in PFMC were also prepared. Concentrations of CRL and surfactants were kept approximately comparable although the technique of DLS using the Zetasizer Nano S is not concentration dependent. Each sample was filtered through the smallest appropriate filter prior to measurement on the instrument at 25 °C. The instrument contains a 4 mW He-Ne laser operating at a wavelength of 633 nm and incorporates non-invasive back scatter optics (NIBS). The measurements were made at a detection angle of 173 ° and the measurement position within the cuvette was automatically determined by the software. Three measurements were taken on each sample to check for reproducibility.
Previously, attempts had been made to measure these samples on an Autosizer 4700 multi-angle instrument fitted with an argon ion laser operating at a wavelength of 488nm. However, the CRL complexes were seen to fluoresce with this laser wavelength and size determination was not possible. The Zetasizer Nano S is fitted a He-Ne laser operating at 633 nm and at this wavelength, fluorescence of the CRL complexes is diminished. This reduction in fluorescence combined with the NIBS optics of the Zetasizer Nano made it possible to accurately determine the size distribution for our enzyme complex.
We report size distribution by volume (nm) with standard deviation in brackets. In this case Z-average and polydispersity index values are not reliable as these samples (particularly surfactants with a molecular weight range) were not monodisperse.
Table 1 summarizes the results obtained from the volume size distribution for CRL, Krytox, KDP and their respective complexes. CRL is a lipophilic enzyme and it is known that it forms aggregates in aqueous solution hence the diameter obtained is far greater than expected for a single molecule of CRL (6.8 nm). Krytox has a larger molecular weight than KDP but is measured at a smaller diameter. It is thought that single molecules of Krytox are being measured here, but it is assumed that KDP is forming micelles and hence a larger diameter is measured. Despite this result, the comparison of CRL Krytox and CRL KDP complexes is highly promising as shown in Figure 2. In the aqueous phase CRL forms aggregates. However on extraction into a fluorous phase it is clear that single enzyme molecules are surrounded by fluorinated surfactant molecules forming a complex as depicted in Figure 1. CRL Krytox is of a slightly larger diameter compared to that for the CRL KDP complex and this is as expected.
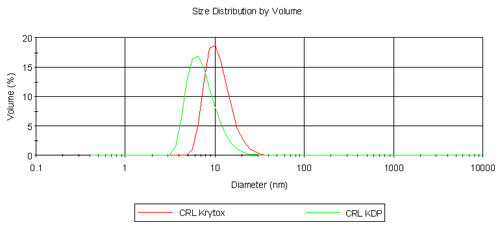
|
Sample | Volume Mean Diameter (nm) |
|---|---|
CRL (aq) | 281.6 (41.1) |
Krytox (fl) | 2.6 (0.1) |
KDP (fl) | 3.9 (1.0) |
CRL Krytox (fl) | 11.0 (0.3) |
CRL KDP (fl) | 7.8 (0.2) |
[1] Horvath I.T. and Rabai J. Science1994, 266, 72
[2] Beier P. and O'Hagan D. Chem. Comm. 2002, 1680
The Zetasizer Nano system from Malvern Instruments is the first commercial instrument to include the hardware and software for combined static, dynamic, and electrophoretic light scattering measurements. The wide range of sample properties available for measurement with the Nano system include, particle size, molecular weight, and zeta potential.
The Zetasizer Nano system was specifically designed to meet the low concentration and sample volume requirements typically associated with pharmaceutical and biomolecular applications, along with the high concentration requirements for colloidal applications. Satisfying this unique mix of requirements was accomplished using a backscatter optical system and a novel cell chamber design. As a consequence of these features, the Zetasizer Nano specifications for sample size and concentration exceed those for any other commercially available dynamic light scattering instrument, with a size range of 0.6nm to 6µm, and a concentration range of 0.1ppm to 40% w/v.
These high sensitivity capabilities can also be applied to real time flow measurements, facilitating Absolute SEC and other HPLC measurements.