Use of the Viscosizer for sizing analysis of Calmodulin samples in different conformational states, and assessment of the technique's complementarity to light scattering in the protein characterization workflow.
Taylor Dispersion Analysis (TDA) with UV-detection as implemented in the Viscosizer instrument, can be used to determine the diffusion coefficient and to calculate the hydrodynamic radius of molecules in solution[1]. This microcapillary flow-based technique uses ultra-low sample consumption, is highly sensitive >100 Daltons, and is compatible with complex solvents or formulations, including those that contain aggregates and/or certain surfactants and excipients which can significantly interfere with light scattering measurements. The instrument baselines on the dispersant itself and uses active referencing, enabling measurement even at very low concentrations. This application note evaluates the sizing analysis of Calmodulin samples by TDA and assesses how this technology can be used to complement light scattering analysis in the protein characterization workflow.
The majority of calcium-binding proteins utilize structural domains known as EF-hands. The EF-hand motif contains two α-helices which are positioned at right angles to each other, separated by an interhelical loop, which forms a single calcium binding site. The ability of these proteins to transport calcium and bind a variety of ligands means they have an important role in many essential processes such as cell signaling, cell motility, growth, proliferation and apoptosis[2]. Their regulatory role in such processes is entirely dependent on the presence or absence of bound calcium ions on the EF-hand regions[3].
Calmodulin (CaM - an abbreviation for 'calcium-modulated protein') is a calcium-binding messenger protein with a number of isoforms, which is abundant in all eukaryotic cells. CaM has a molecular weight of approximately 17kDa, and contains two homologous globular domains with a flexible linker that connects them. Each of these domains contains two EF-hands, which will each bind one Ca2+ ion. Binding of calcium at all four binding sites causes the interhelical angles in the EF-hand motifs to shift, resulting in a "closed" to "open" conformational change. This change exposes hydrophobic interfaces within the protein that are able to bind and subsequently activate a large number of target protein ligands[3].

|
Calmodulin (Sigma Aldrich, Poole, UK) was prepared to a final concentration of 100µg/mL in 10mM HEPES buffer (1M HEPES stock solution (Sigma Aldrich, Poole, UK); Fresenius sterile water (Fresenius Kabi, UK) and adjusted to pH 7 with 0.25M hydrochloric acid and 0.5M sodium hydroxide). Calcium chloride (Sigma Aldrich, Poole, UK) from a 100mM stock CaCl2 solution was added to a 1mL aliquot of the Calmodulin sample, to a final concentration of 0.1mM. Samples for analysis were sterile-syringe filtered (0.02µm (Fisher Scientific, 50-822-149)) and stored at 4ºC until analyzed using the Viscosizer.
All measurements were carried out at 25ºC using default settings, with variable settings adjusted where required. The Viscosizer was fitted with the 214 nm filter unless otherwise stated.
Calmodulin hydrodynamic radius changes according to the presence/absence of calcium chloride
Calmodulin remains in the EF-hand "closed" conformation in the absence of calcium ions, but changes to the EF-hand "open" conformation when all four binding sites are associated with a calcium ion. The estimated hydrodynamic radius (RH) of the "closed" ca. 17kDa globular protein is 2.0nm, with the RH of the protein's "open" linear structure estimated to be 3.23nm[6].
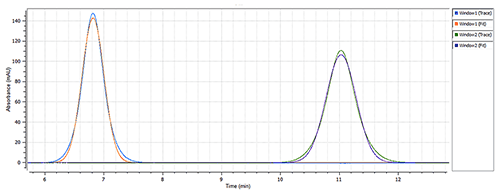
|
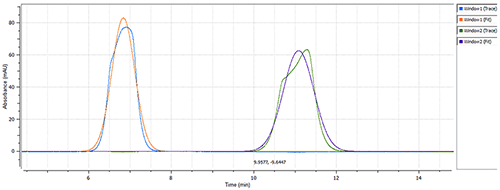
|
The size of Calmodulin was measured at 1.7nm RH (Figure 2) in the "closed" conformation (absence of CaCl2) of the protein (see Figure 1), which is in line with the estimated theoretical value of 2.0nm RH [4]. The presence of 0.1mM calcium chloride caused a significant change in the measured hydrodynamic radius, from 1.7nm to 3.4nm (Figure 3), which agrees with the theoretical size for the "open" conformation of Calmodulin. The same single-component model was used to characterize the data from both data sets, to ensure a robust comparison.
This application note demonstrates that sizing analysis using Taylor Dispersion Analysis (TDA) can be useful as an orthogonal technology to dynamic light scattering (DLS), and explores how the system may be used to monitor the conformational state of proteins in relation to interactions, formulation or stress.