This application note presents the characterization of a potential non-viral gene carrier that has shown transfection ability without significant cytotoxicity in vitro. L-Lysine (C6H14N2O2, abbreviated: Lys or K, 146 g/mol) is an amino acid present in all proteins in the human body.
In this study, a polymer of this amino acid in different pH buffers was investigated. Dendritic poly(L-lysine) of the 6th generation (KG6) is a starburst polypeptide with an expected molar mass of (21+22+23+24+25+26)·146 ~ 18400 g/mol.
Under acidic conditions, the molecule is predicted to be positively charged (the isoelectric point of the single amino acid is ~ 8.9). Figure 1 shows the predicted structure for this molecule at low pH, as modelled by ChemDraw [1].
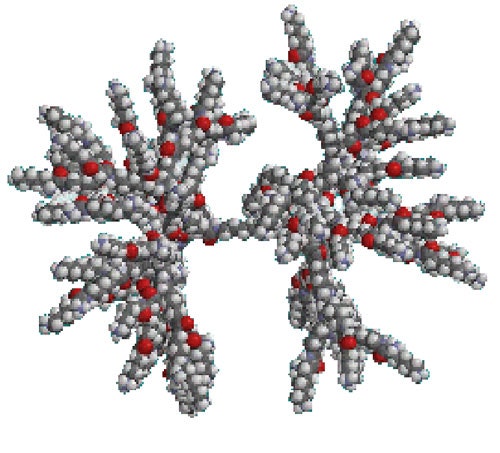
|
When the solvent environment is changed such that the cationic charge neutralizes, the molecule will 'collapse' as the flexible arms no longer need to stretch out. A visual representation of this situation is shown in figure 2, again modelled with ChemDraw.
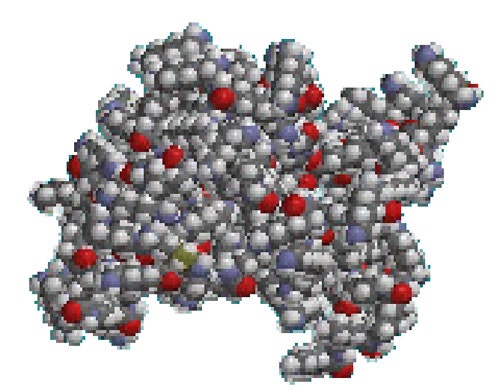
|
KG6 was synthesized in the standard copolymer fashion, and subsequently purified [2]. Samples were dialyzed in different pH buffers (pH 5, 6, 7, and 8) and filtration through a 200nm pore size Anodisk membrane (Whatman).
Dynamic light scattering (DLS) measurements of the samples were made on a Zetasizer Nano S using a detection angle of 173°. All measurements in this study were taken at a temperature of 25°C. The Nano S uses a 4mW He-Ne laser operating at a wavelength of 633nm.
All samples appeared clear and DLS measurements were performed using the automatic settings of the Zetasizer software. Samples were measured at a concentration of 0.5mg/ml
In DLS, the fluctuations of the scattered light are the result of thermal motion of the molecules and these are analyzed by measuring the correlation function of the signal. This leads to the determination of the diffusion coefficient when applying knowledge of the viscosity of the diffusing medium (aqueous buffer in this case).
A distribution of hydrodynamic sizes is obtained based on the intensity of scattered light. This size is the size of an equivalent sphere with the same average diffusion coefficient as the polypeptide molecules. This can be converted into a volume (or mass) size distribution (figure 3).
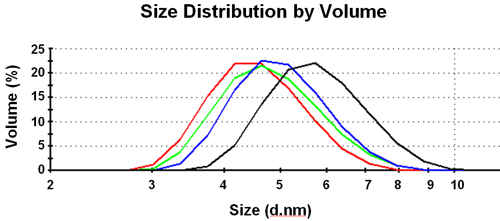
|
At high pH the size is smallest; at low pH the largest mean value for the distribution is observed. The intermediate pH distributions give sizes between these two values. The DLS results are summarized in Table 1.
| Buffer condition | Mean size of Peak (d.nm) | Polydispersity Index (PDI) | MW Estimate, Starburst Polymer (kDa) | MW Estimate, Globular Protein (kDa) |
|---|---|---|---|---|
| pH 5.0 | 5.7 | 0.13 | 21.8 | 38.4 |
pH< 6.0 | 4.9 | 0.14 | 13.8 | 27.9 |
pH 7.0 | 4.7 | 0.14 | 11.7 | 24.9 |
pH 8.0 | 4.5 | 0.15 | 9.9 | 22.1 |
An estimated molecular weight can be obtained from the measured hydrodynamic sizes. The Zetasizer Nano software has several molecular weight estimation models. In this study, the starburst polymer model is compared to the globular protein model.
At pH 5.0, where KG6 is a stiff 'star-polymer', the estimate of 22 kDa is in reasonable agreement with the true 19 kDa. For pH 8.0 on the other hand, KG6 is forming a compact sphere, and the globular protein model provides the better prediction.
The intermediate samples are less well modeled since they neither behave like a dendrimer, nor like a globular compact sphere.
The molecular structure of lysine may be used to predict its charge [3] in solution, shown in figure 4. The charge on the single amino acid is expected to be close to zero at pH 8 leading to a higher influence of van-der-Waals attraction, stronger hydrogen bonds and overall 'shrinkage' of the dendrimer.
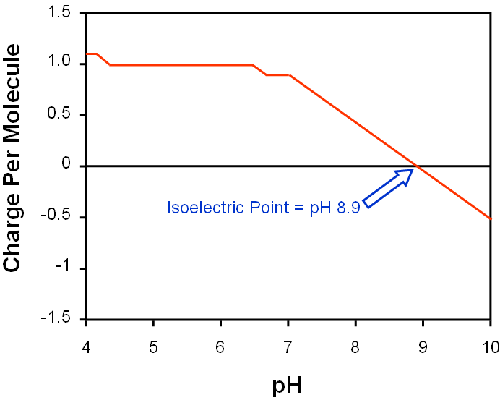
|
A dendritic polypeptide (KG6) was characterized by dynamic light scattering. Results support the existence of a shape change as a function of pH of the buffer. The protein is compact and globular at pH 8, but 'fluffy' and starburst-like at pH 5. This result is expected from the behavior of the individual amino acid L-lysine.
[1] ChemDraw, CambridgeSoft, Massachusetts, USA.
[2] T. Okuda, S. Kawakami, T. Maeie, T. Niidome, F. Yamashita, M. Hashida. J. Controlled Release114(1):69-77 (2006)
[3] http://www.scripps.edu/~cdputnam/ protcalc.html