Antibodies are specialized proteins that function as an essential part of the immune system. They are members of a class of proteins known as immunoglobulins, typically with a molecular weight near 150 kDa. Here, the influence of different storage conditions on the size and quality of these proteins was investigated using light scattering techniques on a Zetasizer Nano S. Dynamic light scattering (DLS) measures the time-dependent fluctuations of the scattered light to measure the diffusion speed and use this to calculate the size of the molecules in solution using the Stokes - Einstein relationship. Static light scattering (SLS) measures the time-averaged intensity to find the molecular weight (MW) of the molecules.
A freshly prepared antibody sample was divided into three portions and stored in different environments:
All samples were prepared at a concentration of 0.8 mg/mL in ammonium hydrocarbonate buffer, at pH=6.8. There was no filtration step in the preparation. The hydrodynamic size of the samples was measured in a 12μL cuvette (ZEN2112) in the Zetasizer Nano S.
A dilution series of each sample was prepared in the same buffer to measure and compare the molecular weight by static light scattering.
All experiments were performed at a temperature of 25°C.
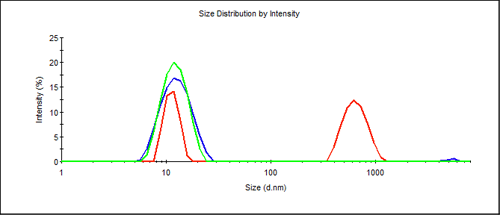
For antibodies the expected size was near 10nm in diameter. Measurements of these antibodies showed a significant difference between the differently treated samples. While all samples showed a peak in the expected range, the proportion of the main peak depended on the conditions (Figure 1). A lower scattering signal was obtained from the protein peak when the sample was stored under non-ideal conditions. It appears that the freeze/thaw cycles reduced the amount of protein present as monomer. In addition, larger aggregates appear after prolonged storage at room temperature or repeated freezing and thawing. The z-average size (overall average of the sample) from a cumulant fit, provides a quick way to identify this sample "quality" through the polydispersity index (PdI) parameter (ISO22412). Table 1 lists data from the three samples.
|
The polydispersity index (PdI) clearly provides a strong indication about whether the sample was harmed by inappropriate storage.
| Storage Condition | Z average (diameter, nm) | PdI |
|---|---|---|
| 35 days at 4°C | 11.4 | 0.075 |
| 31 days at 25°C, then 4°C | 11.7 | 0.172 |
| 5 freeze/thaw cycles, then 4°C | 152 | 0.646 |
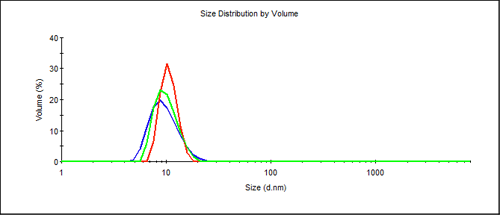
The size distribution from Figure 1 may also be displayed as a volume distribution as shown in Figure 2. The contribution from aggregation is not visible on a standard display, as even in the worst case of the repeatedly frozen sample - the %volume contribution of the main protein peak is still 99.6%. Since light scattering is extremely sensitive to the size of the scattering material it is an ideal technique to find the presence of large components in a protein solution.
|
The observation of the presence of aggregation was confirmed with static light scattering. The expected molecular weight of this sample is about 150 kDa. As shown in Table 2, the results for the 4°C stored sample are in line with the predicted value. A small positive second virial coefficient of 9∙10-4 mL mol / g2 was also obtained, which would indicate that the solution should be stable. The sample stored at room temperature showed a higher molecular weight. This would indicate that part of this sample might be aggregated. Although the repeatedly frozen sample showed the presence of significant aggregation in DLS, the measured molecular weight was very close to that of the monomer. The Zetasizer Nano software's advanced dust rejection algorithm was able to separate the contribution of the large aggregates, and still produced an excellent result.
| Storage Condition | Molecular Weight (kDa) | MW Error (kDa) |
|---|---|---|
| 35 days at 4°C | 150 | +/- 13 |
| 31 days at 25°C, then 4°C | 176 | +/- 6 |
| 5 freeze/thaw cycles, then 4°C | 156 | +/- 12 |
The Zetasizer Nano may be used to evaluate sample stability and quality. Dynamic light scattering is a fast and easy-to-use technique to establish the state of proteins in solution. The most significant sample quality parameter is the polydispersity index (PDI) when comparing effects of storage.