In this application note we describe the use of Nanoparticle Tracking Analysis (NTA) for size and concentration measurements of drug delivery nanoparticles. In addition, by using a fluorescently tagged drug molecule, it was possible to determine how many drug delivery nanoparticles had successfully been loaded with drug molecules.
During the last two decades, biomedical research at a worldwide level has been drastically increasing its efforts to develop nanoparticle based drugs and bring them from the bench to the bedside.
Today, many high-potent molecule-based drugs are used in suboptimal conditions: low dosage (because of their toxicity) or lack of efficiency in the targeting specific organs, cell types, etc.
Thanks to recent discoveries, it has been recognized that nanoparticles, like polymers or liposomes, have excellent properties as drug delivery vehicles to address some of these issues. When considering a nanoparticle-based drug it is critical to characterize the size of these vectors, as it has a considerable effect on their pharmacokinetics or their efficiency and ability to reach their target within the body. Equally, it is also imperative to have an idea of the concentration of nano-objects and more importantly the dosage of nano-objects loaded with the drug of interest.
Here we describe the use of Nanoparticle Tracking Analysis (NTA) for size and concentration measurements of drug delivery nanoparticles. In addition, by using a fluorescently tagged drug molecule, it was possible to determine how many drug delivery nanoparticles had successfully been loaded with drug molecules.
NTA utilizes the properties of both light scattering and Brownian motion in order to obtain the particle size distribution of samples in liquid suspension. A laser beam is passed through the sample chamber, and the particles in suspension in the path of this beam scatter light in such a manner that they can easily be visualized via a 20x magnification microscope onto which is mounted a camera. The camera, which operates at approximately 30 frames per second (fps), captures a video file of the particles moving under Brownian motion within the field of view of approximately 100 μm x 80 μm x 10 μm (Figure A).
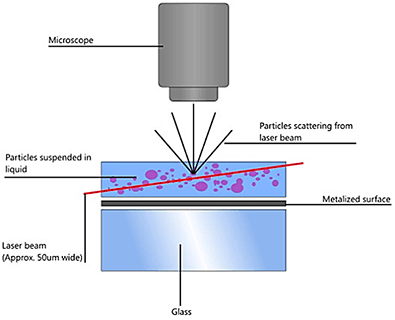
|
The movement of the particles is captured on a frame-by-frame basis. The proprietary NTA software simultaneously identifies and tracks the center of each of the observed particles, and determines the average distance moved by each particle in the x and y planes. This value allows the particle diffusion coefficient (Dt) to be determined from which, if the sample temperature T and solvent viscosity η are known, the sphere-equivalent hydrodynamic diameter, d, of the particles can be identified using the Stokes-Einstein equation (Equation 1).
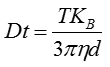
|
NTA is not an ensemble technique interrogating a very large number of particles, but rather each particle is sized individually, irrespective of the others. An example of the size distribution profile generated by NTA is shown in Figure B.
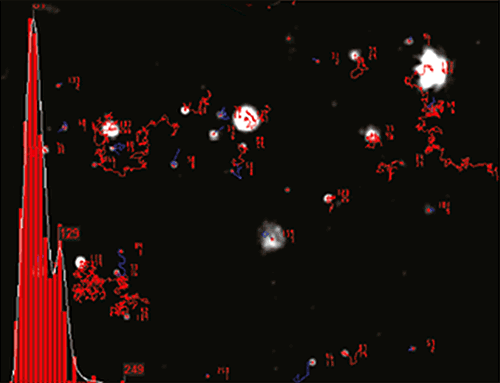
|
In addition, the particles’ movement is measured within a fixed field of view (approximately 100 μm by 80 μm) illuminated by a beam approximately 10 μm in depth. These figures allow a scattering volume of the sample to be estimated; by measuring concentration of the particles within this field of view and extrapolating to a larger volume it is possible to achieve a concentration estimation in terms of particles per mL for any given size class or an overall total. Finally, a fluorescence mode allows differentiation of suitably labelled particles.
A NanoSight LM10 HSBF system was used, comprised of a sCMOS camera, 405 nm laser and 430 nm long-pass filter. Analysis was performed using NTA software 2.3.5.
Polylactic Acid (PLA) particles (Adjuvatis) were designed, synthesized and loaded with a drug of interest conjugated to a fluorescent label (Coumarin6, Excitation 370 nm; Emission 470 mm) (PLA-Coum).
PLA-coum nanoparticles were diluted 100-fold with particle-free ultra-pure water* and mixed by pipetting. * The water was checked by NTA before use and found to be free of contaminant particles.
The diluted sample was loaded into the laser sample chamber using a 1 mL silicon-oil free syringe.
The sample chamber was loaded onto the instrument. The image was adjusted to bring the particles into focus and a video was captured using camera level 7 for 60 seconds to generate data for light-scatter (standard) mode (all particles in the sample measured).With the sample chamber remaining on the instrument, the camera level was increased to maximum (level 16), a second 60 seconds video was taken after insertion of the 430 nm long-pass filter and refocussing to obtain fluorescence-mode data (only particles emitting a fluorescence signal measured).
The videos were then processed using a detection threshold of 4 for both scatter mode and for fluorescent mode. The Min Track Length was set up at 6 for both scatter and fluorescence mode.
Size distribution data were obtained for both measurement modes and overlaid for comparison.
A mixture of labelled and non labelled PLA nanoparticles were expected as a result of the method of sample production. NTA data for these particles both under a scatter mode (Fig.1) and a fluorescence mode (Fig.2) were obtained. The distance moved by each particle from frame to frame of the video was used to generate a diffusion coefficient, and consequently the size of the particle (Fig 1b and Fig 2b), while the average number of particles in each frame of the video was used to give an estimation of sample concentration (Fig 1a and Fig 2a). From these parameters a size distribution profile graph was generated for each measurement mode (Fig 1c and Fig 2c). Summary data for size and concentration are shown in Table 1 and Table 2.
|
|
|
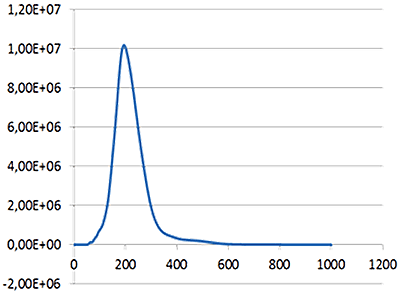
| Mean: | 214 nm |
| Mode: | 199 nm |
| Measured concentration: | 1.23 x 109 particles/mL |
| Dilution factor: | 100 times |
| Final concentration: | 1.23 x 1011 particles/mL |
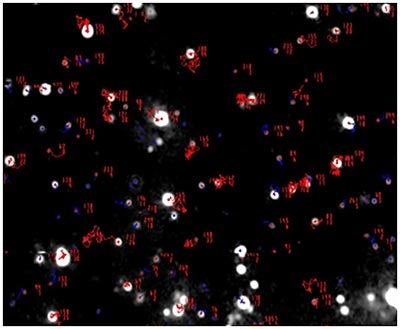
Fig 1a shows the sample viewed under scatter mode. The particles moving under Brownian motion are tracked by NTA 2.3.5 software (Fig.1b) and sample concentration is also estimated. By tracking particle movement from frame to frame in the video, NTA calculates the Diffusion Coefficient for each particle and gives a high resolution size distribution profile (Fig.1c) (x axis = diameter (nm), y axis = concentration ( particles per mL)) Summary size and concentration data are shown in Table 1
|
|
|
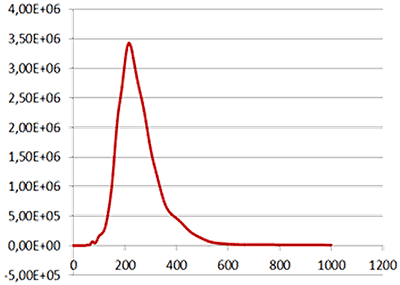
| Mean: | 259 nm |
| Mode: | 223 nm |
| Measured concentration: | 5.31 x 108 particles/mL |
| Dilution factor: | 100 times |
| Final concentration: | 5.31 x 1010 particles/mL |
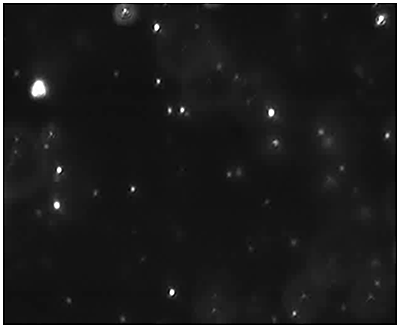
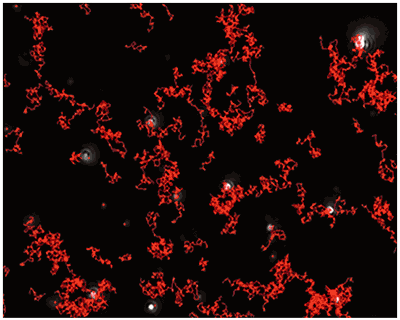
Fig 2a shows the sample viewed under fluorescence mode. With the sample measured through the 430 nm long-pass filter, only the labelled particles are tracked (Fig.2b). Using the Stokes-Einstein equation, particle size is derived from the Diffusion Coefficient obtained from the Brownian motion of the particles. The average number of particles in each frame gives an estimation of particle concentration (Fig2a). The size distribution profile is displayed in Fig2c (x axis = diameter (nm), y axis = concentration (particles per mL)). Summary size and concentration data are shown in Table 2.
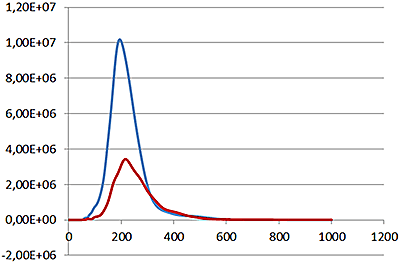
Under scatter mode (no filter), the analysis of the sample (Fig 3 blue line), shows a modal distribution at 199 nm with a final concentration of 1.23e+11 particles/mL. Under fluorescence mode (430 nm long-pass filter inserted), the distribution of the labelled population (Fig 3 red line) shows a slightly bigger mode at 223 nm with a final concentration of 5.31e+10 particles/mL. The widths of the distributions were similar. The sample was also shown to be monodispersed as no subsequent peaks were identified.
These data suggest that the PLA particles are on average potentially 10% larger when loaded with the fluorescently-tagged drug molecule and that over 40% of the drug delivery PLA particles had been successfully loaded with drug molecules.
|
Overlay of the size distribution profiles for PLA-coum nanoparticles in scatter mode (blue line) and fluorescent mode (red line) (x axis = diameter (nm), y axis = concentration (particles per mL)
NanoSight NTA measurements enabled a fast evaluation of the size and concentration of polylactic acid nanoparticles. When loaded with fluorescently labelled drug molecules, the subpopulation of particles containing the fluorophore could be distinguished from the total population giving valuable information on the efficiency of the loading process. These data can be used to identify improved loading processes in a quick and easy manner saving time and effort for drug delivery researchers.