The study of exosomes and other cell-derived microvesicles is an area of rapidly growing importance and the subject of intense interest and research. The previous lack of suitable methods for their detection, analysis, enumeration and phenotyping is proving to be a significant limitation in these studies. This white paper shows the degree to which the technique of Nanoparticle Tracking Analysis (NTA) is helping to address these problems.
NTA utilizes the properties of both light scattering and Brownian motion in order to obtain the particle size distribution of samples in liquid suspension. A laser beam is passed through the sample chamber, and the particles in suspension in the path of this beam scatter light in such a manner that they can easily be visualized via a 20x magnification microscope onto which is mounted a camera. The camera, which operates at approximately 30 frames per second (fps), captures a video file of the particles moving under Brownian motion within the field of view of approximately 100 μm x 80 μm x 10 μm (Figure 1).
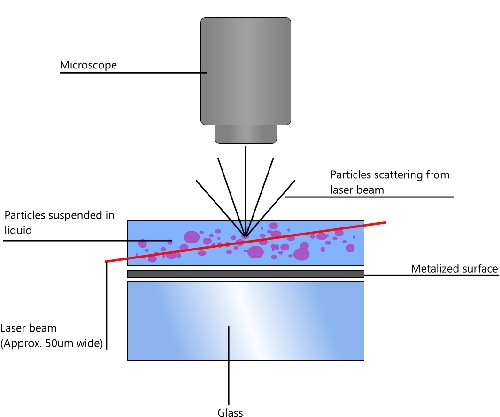
|
The movement of the particles is captured on a frame-by-frame basis. The proprietary NTA software simultaneously identifies and tracks the center of each of the observed particles, and determines the average distance moved by each particle in the x and y planes. This value allows the particle diffusion coefficient (Dt) to be determined from which, if the sample temperature T and solvent viscosity η are known, the sphere-equivalent hydrodynamic diameter, d, of the particles can be identified using the Stokes-Einstein equation (Equation 1).

|
NTA is not an ensemble technique interrogating a very large number of particles, but rather each particle is sized individually, irrespective of the others. An example of the size distribution profile generated by NTA is shown in Figure 2.
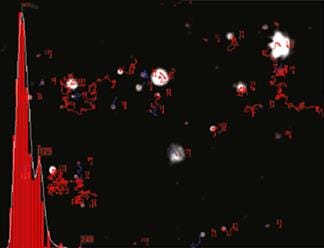
|
In addition, the particles' movement is measured within a fixed field of view (approximately 100 μm by 80 μm) illuminated by a beam approximately 10 μm in depth. These figures allow a scattering volume of the sample to be estimated; by measuring concentration of the particles within this field of view and extrapolating to a larger volume it is possible to achieve a concentration estimation in terms of particles per mL for any given size class or an overall total.
Extracellular vesicular bodies such as microvesicles and exosomes are currently under intense investigation due to their apparently ubiquitous presence in a broad range of prokaryotic and eukaryotic organisms and the wide role they appear to play, at a fundamental level, in many biological processes, both physiological and pathological. Their cellular origin, structure, function and characterization has been extensively reviewed, though still the subject of much debate.
In a recent and comprehensive review, Gyorgy (2011) discussed the technical pitfalls and potential artefacts in the rapidly emerging field, compared results from meta-analyzes of published proteomic studies on membrane vesicles and summarized the clinical implications of membrane vesicles. Following an emerging consensus in terms of nomenclature, he subsequently described exosomes as being 50 -100 nm in diameter and microvesicles as 100 - 1000 nm, and listed those techniques which have been used most frequently in their isolation, purification, detection and analysis (Gyorgy et al. (2011)).
The definition and nomenclature of exosomes and microvesicles is, however, as yet variable. Simpson et al. (2009) define exosomes as 40 -100 nm diameter membrane vesicles of endocytic origin that are released by most cell types upon fusion of multivesicular bodies with the plasma membrane, presumably as a vehicle for cell-free intercellular communication. Because extracellular organelle terminology is often confounding, with many preparations reported in the literature being mixtures of extracellular vesicles, there is a growing need to clarify nomenclature and to improve purification strategies in order to discriminate the biochemical and functional activities of these moieties (Mathivanan et al. (2010)).
Similarly, Lee (2011) also confirmed that because microvesicles (MVs) are so heterogeneous this has led to the usage of multiple names for their designation under different experimental settings. Some of the most frequently encountered descriptors are MVs, microparticles, ectosomes, exosomes, exosome-like vesicles, shed vesicles and most recently oncosomes. Other names have also been used in various specific settings including argosomes, promininosomes, P4 particles, prostasomes, and several others. He stated that to some extent, this diversity reflects the culture of different fields in which MVs have been studied, but also the substantial biological diversity of the underlying biological process (Lee et al. (2011)).
In contrast, platelet-derived microparticles (PMP) are defined as heterogeneous populations of vesicles (<1 μm) generated from the plasma membrane upon platelet activation by various stimulii. They are a discrete population differing from the exosomes which originate from the intracellular multivesicular bodies. PMP also differ from the microparticles derived from megakaryocytes despite the presence of several identical surface markers on the latter. The molecular properties and the functional roles of the PMP are beginning to be elucidated by the rapidly evolving research interest, but novel questions are simultaneously raised (Siljander, 2011).
In conclusion, it is clear that the diversity in nomenclature and definition of microvesicular bodies, be they microvesicles or exosomes, has arisen from the fact that they originate from a very wide range of cellular origins, through a multiplicity of causes and serve multiple functions, all of which are still to be clarified.
Similarly, Herring et al. (2013) focused on the role of cellular exocytic vesiculation in health, disease, and transfusion medicine, recognizing that microparticles (MPs), small membrane-derived vesicles which are derived from many cell types and released into the circulation under shear stress, complement activation, proapoptotic stimulation, cellular damage, or agonist interaction with cell surface receptors.
MVs originate through at least three distinct mechanisms: (a) breakdown of dying cells into apoptotic bodies; (b) blebbing of the cellular plasma membrane (ectosomes); and (c) the endosomal processing and emission of plasma membrane material in the form of exosomes. Their generation may be triggered by pathways involved in oncogenic transformation, microenvironmental stimulation, cellular activation, stress, or death. Vesiculation events occur either at the plasma membrane (ectosomes, shed vesicles) or within endosomal structures (exosomes) (Gyorgy et al. (2011); Lee et al. (2011)).
Exosomes are found in a wide range of bodily fluids such as urine, amniotic fluid, malignant ascites, bronchoalveolar lavage fluid, synovial fluid, breast milk, saliva and blood (Simpson et al. (2009)) and multiple roles have been ascribed to exosomes given the number of different molecular structures associated with their construction. In the case of exosomes derived from breast milk, because exosomes carry immunorelevant structures, they are suggested to participate in directing the immune response and may be important for the development of the infant’s immune system (Admyre et al. (2007)).
A recent patent filing (Chisholm et al. (2013)) concerns the extracellular release of vesicles by photo synthetic cyanobacteria, with NTA used to measure the size distribution of vesicles obtained from the cell-free Prochlorococcus supernatant.
Exosomes are thought to have a significant role in cell signaling and as such exhibit a strong relationship to disease progression. Lee et al. (2011) confirmed that MVs are increasingly recognized as mediators of intercellular communication due to their capacity to merge with, and transfer a repertoire of bioactive molecular content (cargo) to, recipient cells. Such processes may occur both locally and systemically, contributing to the formation of microenvironmental fields and niches. The bioactive cargo of MVs may include growth factors and their receptors, proteases, adhesion molecules, signaling molecules, as well as DNA, mRNA, and microRNA (miRNAs) sequences. As pointed out in numerous studies, the physiological function of exosomes is still a matter of debate, but increasing results in various experimental systems suggest their involvement in multiple biological processes.
More recently, Cicero and Raposo (2012) have reviewed the cell biology of exosomes from an historical perspective and Yuana et al. (2012) have described the tools available to improve the detection of vesicles (including NTA), and the clinical applications being investigated using vesicles for diagnosis, prognosis, and therapy.
As previously discussed, there is an increasing recognition that methods of isolation and preparation of exosomes and microvesicles differ greatly and such differences can have a profound effect on any investigative results obtained. This lack of visibility regarding the true nanoparticulate nature of a sample under study (size, size distribution, number, etc.) has been considered in some detail by Yuana et al. (2011) in their assessment of pre-analytical and analytical issues in the analysis of blood microparticles. They concluded that while results of plasma microparticle (MPs) measurements reported in the literature vary widely, this is clearly not only related to the lack of well standardized MP assays, but also to variations in pre-analytical conditions. Emphasizing the desirability of obtaining fresh platelet-free plasma samples, they also cautioned against inadequate calibration of conventional flow cytometric analysis. When comparing Dynamic Light Scattering (DLS) and NTA, they concluded that the sensitivity of DLS was lower in polydisperse sample types as exemplified by cell-derived MPs. NTA, on the other hand, can accurately size particles in a sample, however larger particles reduce the number of small particles detected by the software. The operation of NTA was not considered, as yet, to be as user friendly as that of DLS, and therefore required some skill in operation. Yuana et al. (2010) had previously found, however, that NTA confirmed the size and number concentration of MPs found by AFM.
The release of exosomes from Epstein-Barr virus transformed B cells has been studied, and NTA (as well as electron microscopy) used to confirm that the nanoparticulate structures observed during these studies were exosomes and not virions attaching to B cells in the samples (Johansson et al. (2010) and Vallhov et al. (2010)). In their study of the potential of exosomes for use in vaccine and immune therapeutic strategies, Vallhov used a number of sophisticated techniques (flow cytometry, confocal laser scanning microscopy, and multispectral imaging flow cytometry) to elucidate interactions with other cell types, but only Electron Microscopy (EM) and NTA were used to discriminate between exosomes and virions in the exosome preparation (Vallhov et al. (2010)).
Similarly, Ludwig and Giebel (2011) used both NTA and EM to size their exosome-enriched solutions, showing they mainly contained particles ranging from 80 to 160 nm, whereas the same sample, when prepared for and documented with EM-based technologies, appears significantly smaller. In a related study, Sokolova et al. (2011) characterized exosomes derived from three different human cell types (HEK 293T, ECFC, MSC) by NTA and SEM and investigated their stability during storage at -20 °C, 4 ºC, and 37 ºC. They showed the size of the exosomes decreased at 4 °C and 37 °C indicating a structural change or degradation. However, neither multiple freezing to -20 °C and thawing, nor multiple ultracentrifugation affected the exosome size. They concluded that NTA was well suited to study exosomes.
Taylor (2011) described the use of NTA for in vivo derived human extracellular vesicles to show sizes 30 to 300 nm. Vesicles at concentrations in the range of 1010 per mL were assessed following chromatographic and affinity isolation of circulating vesicles to identify specific populations of extracellular vesicles.
Gabriel and Giordano (2010) have discussed NTA under the title “Microparticle Sizing and Counting using New Light Scattering Methods” suggesting it offers many advantages to particle size distribution characterization. They suggested that in addition to its ease of operation, speed, and accuracy, the particle size, particle surface characteristics, interaction of the surface with specific ligands, and hydrodynamic volume of the particle are easily obtained. Extensions of these methods also permit the assessment of surface reactions in real time and without reporter group conjugation to the reactant. These methods offer the ability to examine binding constants and kinetics of binding without chemical modification and offer true advantages in product development and clinical diagnostics and therapeutic monitoring.
In describing the use of ultra-filtration (UF), a method which can potentially separate exosomes rapidly based on the characteristics of the physical size, Huang et al. (2012) compared it to more conventional ultra-centrifugation methods. They showed that NTA revealed the size distribution of the main population of particles were from 30 to 150 nm, fitting well to the definition of exosome, suggesting that the UF method is ideal for isolating tumor-associated exosomes from clinical samples. Similar results were showed in other lung cancer cell lines as well as cancer cells and immune cells derived from clinical malignant pleura effusion (MPE) samples. Similarly, Lässer et al. (2012) used NTA in their assessment of a 200 nm filtration before a final 120,000 x g ultracentrifugation as a valuable method of eliminating larger particles, and to evaluate the impact of the filtration step on the RNA profile of the isolated exosome fraction. They concluded that the method used for isolating exosomes affects the RNA profile of the exosome fraction.
Further studies on the use of myristoylated alanine-rich C-kinase substrate (MARCKS) peptide as a probe to target microvesicles (Morton et al. (2012)) employed NTA. It was also used to validate a method for the quantification and profiling of exosomes in human plasma using a protein microarray based on biotin labelled anti-tetraspanin antibodies,CD9, CD63 and CD81 (Jørgensen et al. (2012)), NTA being performed both as total quantification of all microvesicles and with fluorescence-labelling of the exosomes with the detection antibodies).
Soo et al. (2012) established that NTA permitted the determination of both the size distribution and relative concentration of microvesicles, including exosomes, in the supernatants of cultured cells and biological fluids during their study of the release of microvesicles from the human T lymphoblastoid cell lines Jurkat and CEM. They showed that, unstimulated, both cell lines release microvesicles in the size range 70 – 90 nm, which can be depleted from the supernatant by ultracentrifugation at 100,000 x g, and by anti-CD45 magnetic beads, and which (through immunoblotting) also contain the exosome-associated proteins Alix and Tsg101. Incubation with known potentiators of exosome release, the ionophores monensin and A23187, resulted in a significant increase in microvesicle release that was both time and concentration dependent. They concluded that NTA can be effectively applied to monitor microvesicle release from cells of the immune system.
In a study aimed at the setup of a protocol for exosomes isolation from urine, and the quantification and analysis of surface markers and micro-RNA (miRNA) content, Dimuccio et al. (2012) compared and tested four protocols of exosome isolation, based on i) ultracentrifugation (100,000 x g at 4 °C for 1 hour); ii) nanomembrane concentrator Amicon (100k); iii) nanomembrane concentrator Vivaspin 500 (Sartorius); iv) denaturation of Tamm-Horsfall Protein (THP) with DTT (200 mg/mL) followed by ultracentrifugation. Exosome quantification was performed with Bradford assay for protein content, or with NTA concentration measurement. A total mRNA was extracted using mirVana kit (Ambion) and miRNA analysis was performed using quantitative RT-PCR. As exosomes were considered to be smaller than the lower limit of sensitivity of the cytofluorimetric analysis, it was performed after adsorption of isolated vesicles on 4 μm aldehyde–sulphate latex beads. They showed that the protein concentration tested with a Bradford assay only showed a very low exosome concentration for protocol number two, however NTA analysis showed high concentration of exosomes in samples obtained using protocols one and two (4.7 × 108 and 3.5 × 108 exosomes/mL). Their study identified a protocol based on ultracentrifugation as the most suitable to obtain exosomes from urine, in which exosome concentration measurement using NTA was more reliable than protein quantification, possibly due to a contamination by urinary proteins, suggesting their findings could be a valid starting point for the further development of studies in a wide variety of renal pathologies.
Goda et al. (2012) have extended the development of methodologies for the detection of miRNA through the use of a label-free, microelectrode array exploiting the inherent miniaturization of the electrical biosensor meets requirements for massively parallel analysis of circulating microRNA as a non-invasive biomarker. Their study involved the isolation of exosomes from serum-free supernatant of cultured cells by centrifugation, filtration and ultracentrifugation. The isolated exosomes were characterized by NTA.
In their study of the impact of biofluid viscosity on size and sedimentation efficiency of the isolated microvesicles, Momen-Heravi et al. (2012) recognized that the different chemical and molecular compositions of biofluids have an effect on viscosity and this could affect movements of the particles inside the fluid. In addressing the issue of whether viscosity has an effect on sedimentation efficiency of microvesicles using ultracentrifugation they used different biofluids, spiked them with polystyrene beads, and assessed their recovery using NTA to demonstrate that MVs recovery inversely correlates with viscosity. They concluded that, as a result, sample dilutions should be considered prior to ultracentrifugation when processing any biofluids.
Of interest to researchers involved in the isolation, purification and, importantly, storage of exosome samples, Shiba et al. (2012) described their studies on the interaction between the isolated exosomes (from cell culture) and solid materials (including SiO2, Al2O3), and Fang et al. (2012) have highlighted NTA as a promising technique for exosome characterization and quantification in their recent assessments of analytical methods in renal research.
Tatischeff et al. (2012) described the fast characterization of cell-derived extracellular vesicles by NTA, cryo-EM and Raman tweezers microspectroscopy showing that NTA is valuable for studying the size distribution and concentration, Cryo-EM is outstanding for the morphological characterization, including observation of vesicle heterogeneity, while RTM provides the global chemical composition without using any exogenous label. Using cell-derived vesicles of Dictyostelium discoideum, a convenient general model for eukaryotic EVs, pointing out that the size distributions and concentrations of 2 different preparations of D. discoideum EVs obtained after 48 hours of cell growth as measured by NTA were different in terms of size distribution (if not number), meaning that different conditions for the 12,000 x g centrifugation can introduce a biased evaluation of the genuine size distribution of the vesicles in the extracellular medium.Because both cell-culture supernatants and biological fluids contain different types of lipid membranes, it is critical to perform high-quality exosome purification. Théry et al. (2006) described different approaches for exosome purification from various sources, and discussed methods to evaluate the purity and homogeneity of the purified exosome preparations.
Current isolation protocols for their isolation use a two-step differential centrifugation process. Due to their low density, exosomes are expected to remain in the low-speed (17,000 × g) supernatant and to sediment only when the sample is spun at high-speed (200,000 × g). However, other preparation methods have included sucrose gradient centrifugation, Annexin V-coated magnetic beads, immunoisolation, precipitation technologies (ExoQuick®) and filtration technologies (ExoMir®). A typical such isolation and analysis procedure may use a combination of techniques such as that described by Mathias et al. (2009) which employed size filtration followed by ultracentrifugation to isolate and purify exosomes from the colon carcinoma cell line LIM 1215. Morphological visualization and characterization was based on electron microscopy and western blotting, whilst protein identification was achieved using a combination of 1D SDS-PAGE and LC-MS/MS.
However, problems remain. Mathivanan et al. (2010) showed in their recent study on various strategies for purifying exosomes that the transport and propagation of infectious cargo, such as prions, and retroviruses, including HIV (suggesting a role in pathological situations), may be artefacts of exosome-purification strategies. Similarly, Quah and O’Neill (2007) described that exosome fractions of dendritic cells produced in long-term cultures were found to contain Mycoplasma contaminants. The study highlighted the close association between exosomes and infectious agents like Mycoplasma and cautioned about purification procedures for preparation of exosomes for studies on immunity. Furthermore, Bayer-Santos et al. (2012) have shown that the secretion of effector proteins into the extracellular environment by Trypanosoma cruzi is apparently complicated by the fact that T. cruzi releases proteins associated with vesicles that are formed by at least two different mechanisms, as evidenced by proteomic analysis with NTA being used to discriminate different population sizes in parasite conditioned culture supernatant.
Of particular concern in this field is the problem of sample storage and transport, as well of those associated with methods of isolation and purification. Witwer et al. (2013) have investigated anticoagulant, freeze-thaw cycles and RNA isolation methods in EV research as applied to a model of HIV-1 disease. Blood was collected from healthy donors with anticoagulants, including sodium citrate, EDTA, ACD and lithium heparin. Blood was processed immediately to obtain ‘‘platelet-free’’ plasma, and NTA was performed with or without a lipid dye. Quantum dot-conjugated antibodies to surface proteins were also tested. They concluded that NTA indicated minimal effects of anticoagulant and freezing on particle size and number, but specific classes of EVs possibly responded significantly. As expected, some anticoagulants inhibited PCR assays. Newly available biofluids and RNA isolation protocols provided improvements over previous methods; however, RNA extraction should always be optimized carefully.
Sorokina et al. (2013) also undertook the qualitative and quantitative analysis of preservation techniques on extracellular microvesicles in order to facilitate the use of EV for clinical application, recognizing that it is crucial to develop efficient methods for their long-term storage without compromising their function. Quantitative analysis was performed on the EVs pre- and post-preservation (NTA and BCA protein assay) after EVs from lung were stored in PBS (1% DMSO) at 4 and -20 °C for up to 7 days but no differences were seen. Additionally, fresh and preserved EVs did not impact the viability of whole bone marrow cells in co-culture. Fast and efficient methods for isolation of exosomal-like vesicles from cell culture medium and body fluids (urine, saliva, plasma, serum) and exosome isolation, fluorescence labelling, analysis and characterization of cell uptake were also reported in a recent international conference on the subject in Boston USA by Kremenskoy et al. (2013) and Heusermann et al. (2013).
In reviewing the subject of standardization of collection, handling and detection of extracellular vesicles at the same conference, Yuana et al. (2013) pointed out that while the most commonly used method to detect EV is flow cytometry (FCM), it detects only 1-2% of all EV present and accordingly, results from EV research are difficult to compare between laboratories. Thus, they aimed to develop standard collection and handling protocols, and to perform sensitive detection of EV using suitable techniques such as resistive pulse sensing (RPS) and NTA. They found that, in comparison to flow cytometry, RPS and NTA detected 1,000 - 10,000-fold more particles in all EV preparations. In contrast to the above, however, generally the concentration and particle size of EV were more affected by the single freeze/thaw cycle than by centrifugation conditions. Their conclusion was that the type of EV, reconstitution solution and detection limit of techniques used to measure EV are important factors to standardize protocols. The subject of standardization of sample collection, isolation and analysis methods in extracellular vesicle research has also been recently comprehensively reviewed by Witwer et al. (2013). Having been the subject of a recent series of international meetings on the isolation and analysis of EV, purification and analysis of associated RNA molecules, and molecular engineering of EV for therapeutic intervention, it was recognized that there was a clear need for standardization of specimen handling, appropriate normative controls, as well as isolation and analysis techniques to facilitate comparison of results, but also that continual development and evaluation of techniques will be necessary as new knowledge is amassed.
Methods for the isolation and analysis of exosomes and microvesicles has been the subject of much recent patent activity, in which NTA is used as proof of the exosomal nature of the isolates (Vlassov et al. (2013); Antes and Kwei (2013); Jones and Knox, (2013)).
Given the importance and potential role of the RNA cargo carried by microvesicles and exosomes, the development of methods for the extraction and RNA profiling of exosomes has received much attention. Confirming the identity and purity of exosomes by NTA and EM (with Western blots for CD63 marker), Zeringer et al. (2013) aimed to develop protocols for isolation of exosomes from HeLa cell culture media and human blood serum and characterization of their RNA content. Through the use of a “Total exosome RNA and protein isolation kit”, they claimed their isolation procedure was completed in a fraction of the time, compared to the current standard protocols utilizing ultracentrifugation and allowed the recovery of fully intact exosomes in higher yields. Zeringer et al. (2013) then extended this work in a subsequent report in which they used NTA to show their isolation protocol represented a set of reagents and a workflow allowing fast and efficient extraction of exosomes, followed by isolation of RNA and its analysis by qRT-PCR and other techniques.
In an attempt to find out whether spin filtration with size exclusion chromatography (SEC) fractioning might represent a more scalable and reliable method than conventional ultracentrifugation (UC), Nordin et al. (2013) compared UC, spin filtration and spin filtration with sequential LC fractioning for isolation of exosomes from cell culture media. Through RNA and protein content analysis, Western blotting (WB), NTA and electron microscopy, they showed that by simple spin-filtration and sequential LC fractionation, high yields of exosomes can be purified from large media volumes but needed further development to become the gold standard for exosome purification.
An alternative method, involving a novel peptide with affinity for canonical heat shock proteins (HSPs) as a tool for capture and enrichment of extracellular microvesicles (eMV), was proposed by Chute et al. (2013). They showed that eMVs can be purified and evaluated by protein content, and NTA proved comparable to other established methods of eMV isolation. Accordingly, the efficiency of HSP affinity peptide (Vn96)-mediated capture of eMVs matches or exceeds currently accepted methods of eMV isolation while also providing greater specificity in capturing eMVs of particular clinical interest.
Finally, Stensballe et al. (2013) have reported the proteomic analysis of exosomes enriched using exosome microarray.
Admyre C, Johansson SM, Qazi KR, Filén J-J, Lahesmaa R, Norman M, Neve EPA, Scheynius A and Gabrielsson A (2007) Exosomes with Immune Modulatory Features Are Present in Human Breast Milk1 The Journal of Immunology, 2007, 179, 1969 -1978
Antes TJ and Kwei K (2013) Methods For Microvesicle Isolation And Selective Removal, - US Patent 20,130,337,440,
Bayer-Santos E, Aguilar-Bonavides C, Rodrigues SP, Cordero EM, Marques AF, Varela-Ramirez A, Choi H, Yoshida N, da Silveira JF and Almeida IC (2012) Proteomic analysis of Trypanosoma cruzi secretome: characterization of two populations of extracellular vesicles and soluble proteins, Journal of Proteome ResearchJust Accepted Manuscript, DOI: 10.1021/pr300947g, Publication Date (Web): December 8, 2012
Chisholm SW, Biller SJ, Thompson AW (2013) Extracellular release of vesicles by photosynthetic cells, International Patent WO 2013138335 A1,19 Sep 2013
Chute IC,Caissie M., Griffiths S, Chacko S, Davey M, Barnett D, Melville S, Fournier S, Meli MV, Lewis S, Ghosh A and Ouellette RJ (2013) Novel peptide with affinity for canonical heat shock proteins (HSPs) as a tool for capture and enrichment of extracellular microvesicles, Second International Meeting of ISEV 2013, Boston, USA, April 17th-20th, 2013 Journal of Extracellular Vesicles 2013, 2: 20826 - http://dx.doi.org/10.3402/jev.v2i0.20826
Cicero AL and Raposo G (2012) The Cell Biology of Exosomes: Historical and Perspectives, Emerging Concepts of Tumor Exosome–Mediated Cell-Cell Communication, 1-32, DOI: 10.1007/978-1-4614-3697-3_1
Dimuccio V, Ranghino A, Camussi G and Bussolati B (2012) Exosome isolation, count and characterization from normal urine, Nephrol. Dial. Transplant. (2012) 27(suppl 2): ii3-ii4 DOI:10.1093/ndt/gfs196, FO010
Fang DYP, HW King, JYZ Li, JM Gleadle (2012) Exosomes and the Kidney: Blaming the Messenger, - Nephrology, Methods in Renal Research Online ISSN: 1440-1797
Gabriel D A and Giordano K (2010) Microparticle Sizing and Counting Using Light Scattering Methods, Semin Thromb Hemost 36(8): 824-832
Goda T, Masuno K, Nishida J, Kosaka N, Ochiya T, Matsumoto A and Miyahara Y (2012) A label-free electrical detection of exosomal microRNAs using microelectrode array, Chem. Commun., 2012, Advance Article, DOI: 10.1039/C2CC36111F, First published on the web 05 Oct 2012
György B, Szabó T G, Pásztói M, Pál Z, Misják P, Aradi B, László V, Pállinger E, Pap E, Kittel A, Nagy G, Falus A and Buzás E I (2011) Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles, Cell Mol Life Sci.; 68(16): 2667–2688. DOI: 10.1007/s00018-011-0689-3
Herring JM, McMichael MA and Smith SA (2013), Microparticles in Health and Disease. Journal of Veterinary Internal Medicine. doi: 10.1111/jvim.12128
Heusermann W, van Beuran S, Wood M, Morrisey D and Meisner-Kober N (2013) Exosome isolation, fluorescence labelling, analysis and characterisation of cell uptake Second International Meeting of ISEV 2013, Boston, USA, April 17th-20th, 2013 , Journal of Extracellular Vesicles 2013, 2: 20826 - http://dx.doi.org/10.3402/jev.v2i0.20826
Huang W-L, Lin C-C, Su W-C (2012) Isolation of tumor associated exosomes from clinical samples using the ultra-filtration method, International Society for Extracellular Vesicles meeting - ISEV 2012, Gothenburg, Sweden, 18th to 21st April 2012
Johansson S, Vallhov H, Gutzeit C, Li Q, Friend S, George T C, Scheynius A and Gabrielsson S (2010) Exosomes released by Epstein-Barr virus transformed B cells selectively target B cells through CD21-gp350 interactions, International Immunology Meeting Abstracts (2010) 22 (Suppl 1 Pt 4): iv47-iv52. DOI: 10.1093/intimm/dxq251 This article appears in: Day 4: 14th International Congress of Immunology Kobe, Japan
Jones J, Knox SJ (2013) Methods for Fractionation, Analysis and Collection of Microvesicles From Patient Samples, - US Patent 20,130,095,575, 2013
Jørgensen M, Bæk R, Søndergaard E, Pedersen S, Kristensen S R, and Varming K(2012) Quantification and Profiling of Exosomes in Human Plasma using Protein Microarray, International Society for Extracellular Vesicles meeting - ISEV 2012, Gothenburg, Sweden, 18th to 21st April 2012
Jørgensen M, Bæk R., Søndergaard EKL, Pedersen S, Kristensen SR and Varming K (2013) Antigenic capturing, detection and phenotyping of exosomes using protein microarray, Second International Meeting of ISEV 2013, Boston, USA, April 17th-20th, 2013 , Journal of Extracellular Vesicles 2013, 2: 20826 - http://dx.doi.org/10.3402/jev.v2i0.20826
Kremenskoy M, Kremenska Y, Kurochkin I (2013) Fast and Efficient Method for isolation of exosomal-like vesicles from cell culture medium and body fluids; Urine, saliva, plasma, serum, Second International Meeting of ISEV 2013, Boston, USA, April 17th-20th, 2013 , Journal of Extracellular Vesicles 2013, 2: 20826 - http://dx.doi.org/10.3402/jev.v2i0.20826
Lässer C, Sjöstrand M and Lötvall J (2012) The pellet and the pellet cap of unfiltered extracellular vesicles have different characteristics and RNA content, International Society for Extracellular Vesicles meeting - ISEV 2012, Gothenburg, Sweden, 18th to 21st April 2012
Lee T H, D’Asti E, Magnus N, Al-Nedawi K, Meehan B and Rak J (2011) Microvesicles as mediators of intercellular communication in cancer—the emerging science of cellular ‘debris’, Semin Immunopathol, DOI 10.1007/s00281-011-0250-3
Ludwig A-K and Giebel B (2011) Organelles in focus, Exosomes: Small vesicles participating in intercellular communication, The International Journal of Biochemistry & Cell Biology, In Press, DOI:10.1016/j.biocel.2011.10.005
Mathias RA, Lim JW, Ji H and Simpson RJ (2009) Isolation of Extracellular Membranous Vesicles for Proteomic Analysis, in Membrane Proteomics: Methods and Protocols, Methods in Molecular Biology, Vol 528, p 227-242
Mathivanan S, JH, Simpson RJ (2010) Exosomes: extracellular organelles important in intercellular communication J Proteomics. 2010 Sep 10;73(10):1907-20. Epub 2010 Jul 1.
McAlexander MA, Ihms EA, Muth D, Sangal N, Metcalf Pate KA and Witwer KW (2013) Investigations of freeze-thaw cycles, anticoagulant and RNA isolation methods in biofluids research and application of methods to a model of HIV-1 infection Second International Meeting of ISEV 2013, Boston, USA, April 17th-20th, 2013 , Journal of Extracellular Vesicles 2013, 2: 20826 - http://dx.doi.org/10.3402/jev.v2i0.20826
Momen-Heravi F, Balaj L, Alian S, Trachtenberg A J, Hochberg F H, Skog J, and Kuo W P (2012) Impact of Biofluid Viscosity on Size and Sedimentation Efficiency of the Isolated Microvesicles, Front Physiol. 2012; 3: 162
Morton L, Saludes J, Beninson L, Chapman E, Fleshner M, Hang Y (2012) Marcks Peptide As A Probe To Target Microvesicles, International Society for Extracellular Vesicles meeting - ISEV 2012, Gothenburg, Sweden, 18th to 21st April 2012
Nordin JZ, Lee Y, Wiklander OPB, Vader P, Smith CIE and EL Andaloussi S (2013) LC purification of exosomes: way forward or dead end?, Second International Meeting of ISEV 2013, Boston, USA, April 17th-20th, 2013 , Journal of Extracellular Vesicles 2013, 2: 20826 - http://dx.doi.org/10.3402/jev.v2i0.20826
Quah BJC. and O’Neill HC.(2007) Mycoplasma contaminants present in exosome preparations induce polyclonal B cell responses, Journal of Leukocyte Biology.;82:1070-1082.
Shiba K and Suga K (2012) Interaction between exosomes and surfaces of materials, International Society for Extracellular Vesicles meeting - ISEV 2012, Gothenburg, Sweden, 18th to 21st April 2012
Siljander PRM (2011) Platelet-derived microparticles – an updated perspective, Thrombosis Research, Volume 127, Supplement 2, January 2011, Pages S30-S33, Proceedings of the 43rd Nordic Coagulation Meeting - Interaction between clinic and laboratory
Simpson RJ, Lim JW, Moritz RL, Mathivanan S (2009) Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics. 2009 Jun;6(3):267-83.
Sokolova V, Ludwig A.-K, Hornung S, Rotan O, Horn P A , Epple M and Giebel B. (2011) Characterisation of exosomes derived from human cells by Nanoparticle Tracking Analysis and scanning electron microscopy, Colloids and Surfaces, B: Biointerfaces, DOI:10.1016/ j.colsurfb.2011.05.013.
Soo CY, Song Y, Zheng Y, Campbell E C, Riches AC, Gunn-Moore F and Powis SJ (2012) Nanoparticle Tracking Analysis monitors microvesicle and exosome secretion from immune cells. Immunology, 'Accepted Article', DOI: 10.1111/j.1365-2567.2012.03569.x
Sorokina A, Aliotta JM, Pereira M, Dooner MS, Wen S, Goldberg LR, Adler A, DelTatto M, Papa E, Quesenberry PJ (2013) Qualititative and quantitative analysis of preservation techniques on extracellular microvesicles Second International Meeting of ISEV 2013, Boston, USA, April 17th-20th, 2013 , Journal of Extracellular Vesicles 2013, 2: 20826 - http://dx.doi.org/10.3402/jev.v2i0.20826
Stensballe A, Jorgensen M, Baek R, Pedersen S & Varming K (2013) Proteomic analysis of exosomes enriched using exosome microarray, Second International Meeting of ISEV 2013, Boston, USA, April 17th-20th, 2013 , Journal of Extracellular Vesicles 2013, 2: 20826 - http://dx.doi.org/10.3402/jev.v2i0.20826
Tatischeff I, Larquet E, Falcón-Pérez J, Turpin P & Kruglik S (2012). Fast characterisation of cell-derived extracellular vesicles by nanoparticles tracking analysis, cryo-electron microscopy, and Raman tweezers microspectroscopy. Journal Of Extracellular Vesicles, 1. Retrieved from http://www.journalofextracellularvesicles.net/index.php/jev/article/view/19179/24812
Taylor DD (2011) Nanoparticle tracking analyses of in vivo derived human extracellular vesicles Exosomes and Microvesicles 2011, Wyndham Hotel, Lake Buena Vista, Florida October 15 - 17, 2011
Théry C, Amigorena S, Raposo G and Clayton A (2006). Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids. Current Protocols in Cell Biology. 30:3.22.1–3.22.29
Vallhov H, Gutzeit C, Johansson S M, Nagy N, Paul M, Li Q, Friend S, George T C, Klein E, Scheynius A and Gabrielsson S (2010) Exosomes Containing Glycoprotein 350 Released by EBV-Transformed B Cells Selectively Target B Cells through CD21 and Block EBV Infection In vitro The Journal of Immunology November 24, 2010 DOI: 10.4049/jimmunol.1001145
Vlassov A, Li M, Zeringer E, Conrad R (2013) Methods And Compositions For Exosome Isolation, - US Patent 20,130,273,544, 2013
Witwer KW., Buzás EI, Bemis LT, Bora A, Lässer C, Lötvall J, Nolte-‘t Hoen EN, Piper MG, Sivaraman S, Skog J, Théry C, Wauben MH, Hochberg F (2013) Standardization of sample collection, isolation and analysis methods in extracellular vesicle research, Journal of Extracellular Vesicles 2013, 2: 20360 - http://dx.doi.org/10.3402/jev.v2i0.20360
Witwer K, Muth D, Sangal N and McAlexander M. (2013) Investigations of anticoagulant, freeze_thaw cycles and RNA isolation methods in EV research and application of methods to a model of HIV-1 disease, Second International Meeting of ISEV 2013, Boston, USA, April 17th-20th, 2013 , Journal of Extracellular Vesicles 2013, 2: 20826 - http://dx.doi.org/10.3402/jev.v2i0.20826
Yuana Y, Oosterkamp TH, Bahatyrova S, Ashcroft B, Rodriguez PG, Bertina R M and Osanto S (2010) Atomic force microscopy: a novel approach to the detection of nanosized blood microparticles, Journal of Thrombosis and Haemostasis, Volume 8 Issue 2, Pages 315 - 323
Yuana Y; Bertina R M; Osanto S (2011) Pre-analytical and analytical issues in the analysis of blood, Microparticles, Thrombosis and Haemostasis, 105.3
Yuana Y, Sturk A, Nieuwland R (2012) Extracellular vesicles in physiological and pathological conditions, Blood Reviews, Available online 20 December 2012, http://dx.doi.org/10.1016/j.blre.2012.12.002
Yuana Y., Boing AN, Hau CM, Grootemaat AE, Sturk A and Nieuwland R (2013) Standardisation of collection, handling and detection of extracellular vesicles, Second International Meeting of ISEV 2013, Boston, USA, April 17th-20th, 2013 , Journal of Extracellular Vesicles 2013, 2: 20826 - http://dx.doi.org/10.3402/jev.v2i0.20826
Zeringer E, Li M, Barta T, Schageman J, Pedersen KW, Neurauter A, Magdaleno S, Setterquist R and Vlassov AV (2013) Methods for the extraction and RNA profiling of exosomes, World J Methodol 2013 March 26; 3(1): 11-18