Confident characterization of protein stability
Differential Scanning Calorimetry (DSC) is an analysis technique used to characterize the stability of a protein or other biomolecule directly in its native form. It does this by measuring the heat change associated with the molecule’s thermal denaturation when heated at a constant rate.
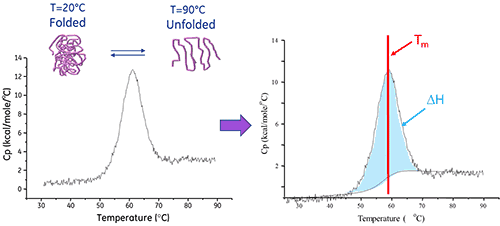
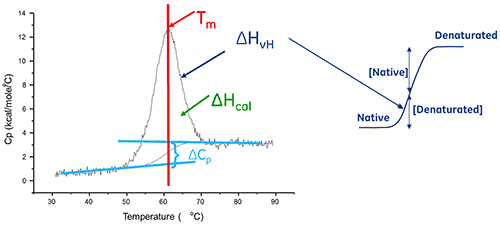
A biomolecule in solution is in equilibrium between its native (folded) and denatured (unfolded) conformations. The higher the thermal transition midpoint (Tm), the more stable the molecule. DSC measures the enthalpy (∆H) of unfolding that results from heat-induced denaturation. It is also used to determine the change in heat capacity (ΔCp) of denaturation. DSC can elucidate the factors that contribute to the folding and stability of native biomolecules. These include hydrophobic interactions, hydrogen bonding, conformational entropy and the physical environment.
The precise and high quality data obtained from DSC provides vital information on protein stability in process development, and in the formulation of potential therapeutic candidates.
Macromolecules and macromolecular assemblies (>5000 Daltons), such as proteins, nucleic acids and lipids, can form well-defined structures that undergo thermally-induced conformational changes. These structural rearrangements result in the absorption of heat caused by the redistribution of non-covalent bonds. Differential scanning calorimeters measure this heat uptake.
The thermal core of a DSC system consists of two cells, a reference and a sample cell.
The device is designed to maintain the two cells at the same temperature, as they are heated.
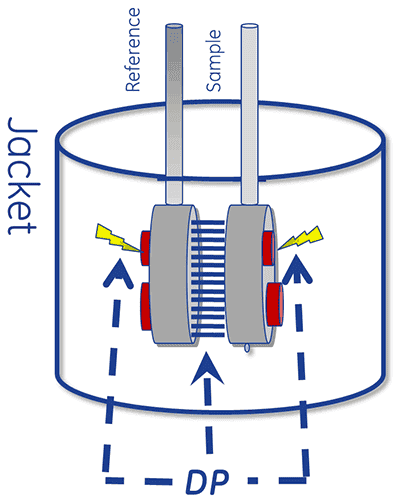
To perform a DSC measurement, the reference cell is first filled with buffer and the sample cell with the sample solution. These are then heated at a constant scan rate.
The absorption of heat that occurs when a protein unfolds causes a temperature difference (ΔT) between the cells, resulting in a thermal gradient across the Peltier units. This sets up a voltage, which is converted into power and is used control the Peltier to return ΔT (the temperature differential) to 0°C. Alternatively, the cells can be allowed to reach thermal equilibrium passively through conduction.
The enthalpy of protein unfolding is the area under the concentration-normalized DSC peak and has units of calories (or joules) per mole.
In certain cases, thermodynamic models can be fitted to the data to obtain the Gibb's free energy (ΔG), the calorimetric enthalpy (ΔHcal), the van't Hoff enthalpy (ΔHvH), the entropy (ΔS) and the change in the heat capacity (ΔCp) associated with the transition.
Differential scanning calorimetry analysis is widely used for drug discovery and development. Key applications include:

MicroCal PEAQ-DSC AutomatedGold standard protein stability analysis for the regulated environment |

MicroCal PEAQ-DSCGold standard protein stability analysis for research applications |
|
|---|---|---|
| Measurement type | ||
| Label-free analysis | ||
| Protein stability | ||
| Technology | ||
| Differential Scanning Calorimetry (DSC) | ||
| Sample throughput | Up to - 50per 24h day | Up to - 6per 8h day |
| Temperature range | 2°C - 130°C | 2°C - 130°C |