Features and benefits
MicroCal PEAQ-ITC is designed for ease of use and exceptional sensitivity. The system directly measures the heat released or absorbed during biochemical binding events, from which it calculates binding affinity (KD), stoichiometry (n), enthalpy (ΔH), and entropy (ΔS).
With minimal sample preparation and system optimization data is generated quickly and easily. The wide affinity range enables analysis of weak to high affinity binders, with excellent reproducibility.
MicroCal ITC is an essential tool for any research laboratory studying biomolecular interactions where high sensitivity and fast results are paramount.
- User-friendly guided workflows with embedded help videos give any level of user the ability to generate high quality data
- High signal to noise gives more confidence in accessing data quality and relevance of generated affinity and thermodynamic parameters
- Automated washing with detergent of the sample cell and titration syringe assists in producing high quality reproducible data
- All binding parameters (affinity, stoichiometry, enthalpy and entropy) in a single experiment
- Quick to first results with no assay development, no labeling, no immobilization and no molecular weight limitations
- Sensitivity to investigate any biomolecular interaction using as little as 10µg of protein
- Directly measures millimolar to nanomolar affinities (KDS) (10-2 to 10-9 M)
- Measures nanomolar to picomolar disassociation constants using competitive binding techniques (10-9 to 10-12M)
- Nonreactive Hastelloy for chemical resistance and compatibility with biological samples
- Compatible with non-aqueous solvents
- Upgradable to fully automated MicroCal PEAQ-ITC Automated
A wide range of applications can be investigated with MicroCal PEAQ-ITC, including the characterization of molecular interactions of small molecules, proteins, antibodies, nucleic acids, lipids and other biomolecules. It can also be used to measure enzyme kinetics.
Analysis Software
MicroCal PEAQ-ITC analysis software offers experiment design simulation, batch evaluation of large data sets, automated assessment of data quality and a streamlined user interface that guides the user to final figures and presentation quality graphs quickly and easily.
- Open multiple experiments in a single session
- Supports many automated fitting models
- Dissociation
- Enzyme Kinetics - Multiple Injections
- Enzyme Kinetics - Single Injection
- One Set of Sites
- Sequential Binding Sites
- One Set of Sites - SIM
- Two Sets of Sites
- Competitive
- Automated assessment of data quality
- Good quality data – Binding
- Good quality data - No binding
- Poor quality data – Check data
How ITC works
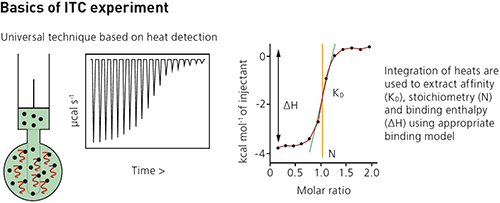
Isothermal titration microcalorimeters measure the heat change that occurs when two molecules interact.
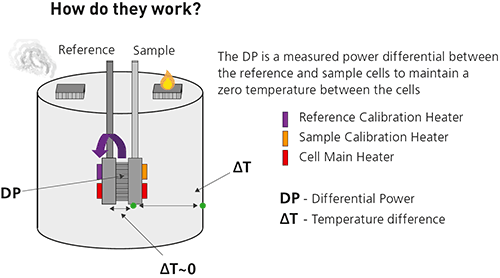
Heat is released or absorbed as a result of the redistribution and formation of non-covalent bonds when the interacting molecules go from the free to the bound state. ITC monitors these heat changes by measuring the differential power, applied to the cell heaters, required to maintain zero temperature difference between the reference and sample cells as the binding partners are mixed.
The reference cell usually contains water, while the sample cell contains one of the binding partners (the sample, often but not necessarily a macromolecule) and a stirring syringe which holds the other binding partner (the ligand). The ligand is injected into the sample cell, typically in 0.5 to 2 μL aliquots, until the ligand concentration is two- to three-fold greater than the sample.
Each injection of ligand results in a heat pulse that is integrated with respect to time and normalized for concentration to generate a titration curve of kcal/mol vs molar ratio (ligand/sample). The resulting isotherm is fitted to a binding model to generate the affinity (KD), stoichiometry (n) and enthalpy of interaction (ΔH).