Using Insulin as a case study, Taylor Dispersion measurements are used to monitor self-association and oligomeric state in a variety of buffer conditions, and have the ability to render surfactants and excipients invisible in measurement data.
Taylor Dispersion Analysis (TDA) is an orthogonal technique for sizing and stability studies of biomolecules in solution. Studies have shown that hydrodynamic radius (Rh) measurements from TDA on monodisperse systems such as monoclonal antibodies (mAbs), are comparable to data obtained using the well-established and complementary technique of Dynamic Light Scattering (DLS)1.
Protein stability is one of the primary concerns during biopharmaceutical development, as it negatively impacts upon immunogenicity and potency of the biotherapeutic. Screening of candidate molecules for developability under a variety formulation conditions is therefore a critical aspect in early stages of the development pipeline.
It is important to monitor any potentially adverse effects that buffer matrices, excipients and surfactants may have on the molecule of interest, such as changes in conformational stability and self-association. However observing changes occurring specifically to the target molecule can be analytically difficult, complicated by the complex background of the excipients themselves.
Notably for protein stability assessment under formulation conditions, the use of TDA with a matched sample buffer effectively renders excipients and surfactants invisible, enabling label-free characterization of target biomolecules in complex solutions. This is in contrast to light scattering measurements for example, where data can become more and more dominated by scattering interference from excipients in the formulation2.
As an important biotherapeutic used in the treatment of Diabetes Mellitus, Insulin provides an interesting case study for the use of TDA as a technique for characterizing the hydrodynamic radius (Rh) and self-association and stability of protein-based biopharmaceuticals in complex solutions.
The self-association of Insulin is fundamental to the protein’s function within the body; hexameric Insulin is the more stable form used to store the protein within cells, whereas monomeric Insulin is the active form that binds to Insulin receptors3. Identifying the oligomeric state of Insulin in a specific formulation is therefore important in predicting the stability of the formulation. Hexamers are formed through the self-association of Insulin monomers into dimers, which combine to form hexamers. Changes in Insulin’s oligomeric state can be observed as a change in size, where smaller sizes correspond to lower order oligomers.
Changes to environment can cause changes to the protein itself, and Insulin can be found in a number of different oligomeric states under different formulation conditions.
In this study, Taylor Dispersion Analysis (TDA) has been used to assess the oligomeric state and stability of Insulin under varying buffer and excipient conditions.
TDA is a microcapillary flow-based technique, whereby a nanoliter volume sample pulse is injected into a laminar flow of matched sample buffer. Analysis of the time-evolved concentration profile (or Taylorgram) from UV absorption allows the molecular diffusion coefficient and hence hydrodynamic radius of solute molecules to be determined4.
Figure 1 shows the variation in self-association and oligomeric state of Insulin in a selection of different formulation buffers as measured by TDA. These data follow the same trends that are well characterized within the literature5.
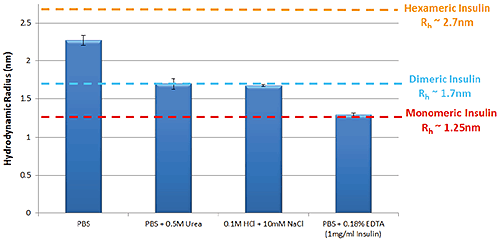
Figure 1: Hydrodynamic radius of Insulin measured by TDA in a range of buffer conditions. Insulin concentration is 2mg/ml (unless stated)
Arginine is an excipient added to formulations in order to improve thermal stability6. Figure 2 is a thermal trend plot measured using DLS (Zetasizer Nano), showing how the addition of Arginine increases the temperature at which Insulin aggregates.
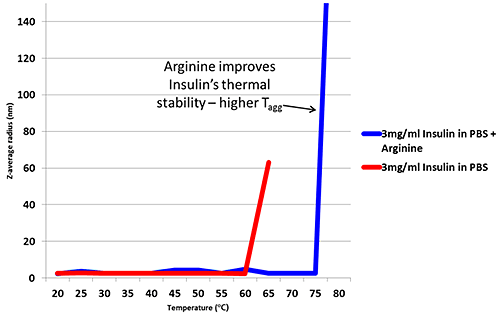
Figure 2: Thermal trend measured by DLS (Zetasizer Nano) showing an increased aggregation temperature for Insulin in PBS with the addition of Arginine
DLS provides a useful tool for monitoring thermal stability, but when measuring changes to a molecule of interest, light scattering results can be influenced by excipients particularly when the excipient is of similar size to the target biomolecule.
Figure 3 shows how the Arginine present in the buffer is picked up as a separate intensity peak in DLS, whereas the TDA Taylorgram is not affected by the addition of excipients. TDA is able to selectively monitor the molecule of interest, independent of formulation components.
Note the Malvern Viscosizer uses twin window detection methodology for TDA, hence the two peaks shown in the Taylorgrams in Figure 3 are two measurements from the same sample pulse that are temporally-separated at different detection points along the microcapillary.
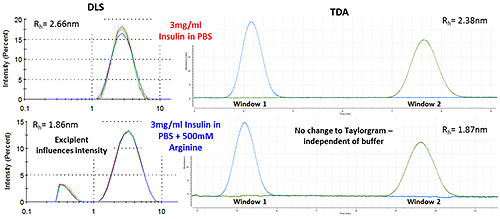
Figure 3: DLS (Intensity vs size plots) and TDA Taylorgrams (UV Absorbance vs time plots) for solutions of 3mg/ml Insulin in PBS with and without the addition of Arginine.
The influence of formulation components such as excipients and surfactants on protein stability and self-association in solution is important to understand, as biotherapeutic products generally require complex formulations.
Light scattering techniques are well established as a screen for early stage developability, but the influence of complex mixtures of excipients and surfactants in the formulation matrix can interfere significantly with measurement data, from reducing measurement sensitivity (particularly at low protein concentrations) to masking the response from the target protein molecule.
Using Insulin as a case study, this Application Note demonstrates how Taylor Dispersion Analysis can be used to monitor self-association and oligomeric state for a variety of buffer conditions.
Notably, the use of specific UV wavelength detection and matched sample buffer effectively tunes TDA to be selective for the target protein molecule, and renders the formulation components invisible in measurement data.
TDA is particularly well suited as an orthogonal technique for early stage developability screening for biotherapeutics, especially as this automated technique is applicable at low concentration with ultra-low sample volume requirements.